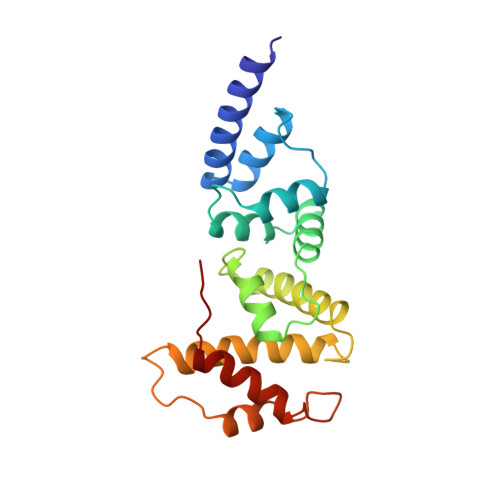
Structure of the Sec7 domain of the Arf exchange factor ARNO.
Cherfils, J., Menetrey, J., Mathieu, M., Le Bras, G., Robineau, S., Beraud-Dufour, S., Antonny, B., Chardin, P.(1998) Nature 392: 101-105
- PubMed: 9510256
- DOI: https://doi.org/10.1038/32210
- Primary Citation of Related Structures:
1PBV - PubMed Abstract:
Small G proteins switch from a resting, GDP-bound state to an active, GTP-bound state. As spontaneous GDP release is slow, guanine-nucleotide-exchange factors (GEFs) are required to promote fast activation of small G proteins through replacement of GDP with GTP in vivo. Families of GEFs with no sequence similarity to other GEF families have now been assigned to most families of small G proteins. In the case of the small G protein Arf1, the exchange of bound GDP for GTP promotes the coating of secretory vesicles in Golgi traffic. An exchange factor for human Arf1, ARNO, and two closely related proteins, named cytohesin 1 and GPS1, have been identified. These three proteins are modular proteins with an amino-terminal coiled-coil, a central Sec7-like domain and a carboxy-terminal pleckstrin homology domain. The Sec7 domain contains the exchange-factor activity. It was first found in Sec7, a yeast protein involved in secretion, and is present in several other proteins, including the yeast exchange factors for Arf, Geal and Gea2. Here we report the crystal structure of the Sec7 domain of human ARNO at 2 A resolution and the identification of the site of interaction of ARNO with Arf.
Organizational Affiliation:
Laboratoire d'Enzymologie et Biochimie Structurales, CNRS UPR 9063, Gif-sur-Yvette, France. cherfils@lebs.cnrs-gif.fr