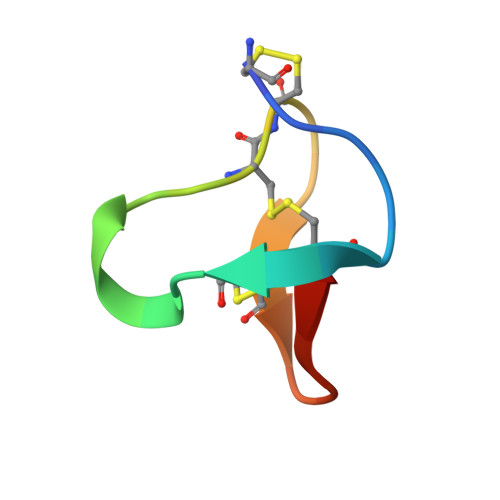
Solution structure of omega-conotoxin MVIIC, a high affinity ligand of P-type calcium channels, using 1H NMR spectroscopy and complete relaxation matrix analysis.
Farr-Jones, S., Miljanich, G.P., Nadasdi, L., Ramachandran, J., Basus, V.J.(1995) J Mol Biol 248: 106-124
- PubMed: 7731037
- DOI: https://doi.org/10.1006/jmbi.1995.0205
- Primary Citation of Related Structures:
1OMN - PubMed Abstract:
We have determined the solution structure of the omega-conotoxin MVIIC from Conus magus by 1H NMR. This conopeptide preferentially blocks P and Q type Ca2+ currents by binding with high affinity to voltage-sensitive Ca2+ channels in neurons. This 26 residue peptide with three disulfide bonds was chemically synthesized and refolded for NMR structural studies. The 1H NMR NOESY spectrum of this peptide was completely assigned, with stereospecific assignments made for 12 of the beta prochiral centers. Complete relaxation matrix analysis using MARDIGRAS was used to obtain initial interproton distances from peak intensities. The correlation time necessary for these calculations was determined by measuring 13C relaxation times using inversely detected natural abundance spectra. Distances were input to DG, which provided 15 starting structures which were then subjected to restrained molecular dynamics calculations using SANDER with the AMBER 91 force field in vacuo. 1H-1H vicinal coupling constants were obtained using a combination of line fitting of both E. COSY and NOESY spectra and used to generate angle restraints that were included explicitly in the restrained molecular dynamics calculations. The final set of the 15 best structures had a backbone rmsd of 0.84 A. The ensemble R1/6 factor calculated by CORMA for the final 15 structures was 11%. The final structure consists of an anti-parallel, triple-stranded beta-sheet, with four turns. In spite of significant differences in amino acid sequence and affinities for calcium channel subtypes, the backbone structure of omega-conotoxin MVIIC is very similar to the previously reported structure of omega-conotoxin GVIA.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California, San Francisco 94143-0446, USA.