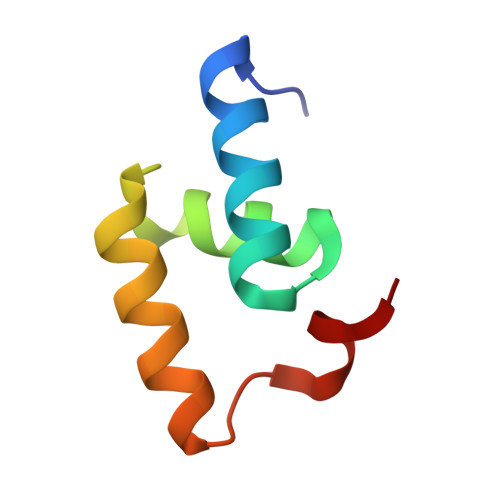
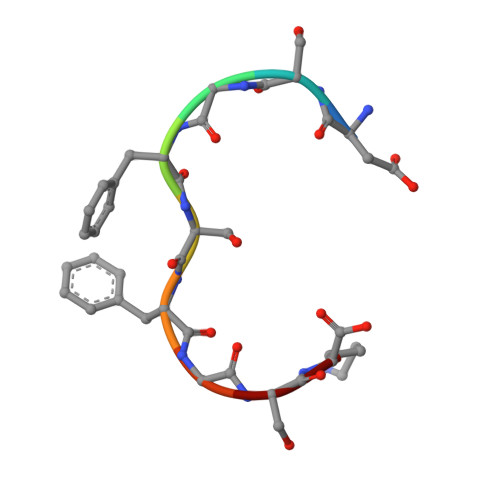
Structural Basis for the Interaction between the Tap/Nxf1 Uba Domain and Fg Nucleoporins at 1 A Resolution
Grant, R.P., Neuhaus, D., Stewart, M.(2003) J Mol Biol 326: 849
- PubMed: 12581645
- DOI: https://doi.org/10.1016/s0022-2836(02)01474-2
- Primary Citation of Related Structures:
1OAI - PubMed Abstract:
The mRNA nuclear export function of Tap/NXF1 requires interactions with nuclear pore proteins (nucleoporins) that contain characteristic Phe-Gly repeats based on FG, GLFG or FxFG cores separated by hydrophilic linkers. FG-nucleoporins bind the two most C-terminal domains of Tap, which have NTF2 and UBA folds, respectively. We used a combination of NMR and X-ray crystallography to define the interaction interface between Tap UBA and FxFG nucleoporins and show that it involves primarily the two aromatic rings of the FxFG core that bind in a hydrophobic surface depression centred on Tap Cys588. NMR evidence indicates that the same depression mediates the binding of GLFG nucleoporins, which we confirmed by demonstrating competition between the two classes of repeat for binding to Tap UBA. Moreover, modification of Cys588 reduced the binding of Tap UBA to both GLFG and FxFG nucleoporins as well as to nuclear envelopes. These data underscore the central role of the conserved FG-nucleoporin repeat cores in binding to Tap UBA and indicate that functional differences between different classes of nucleoporins depend more on their spatial distribution in nuclear pores than on their binding to different sites on Tap UBA.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK.