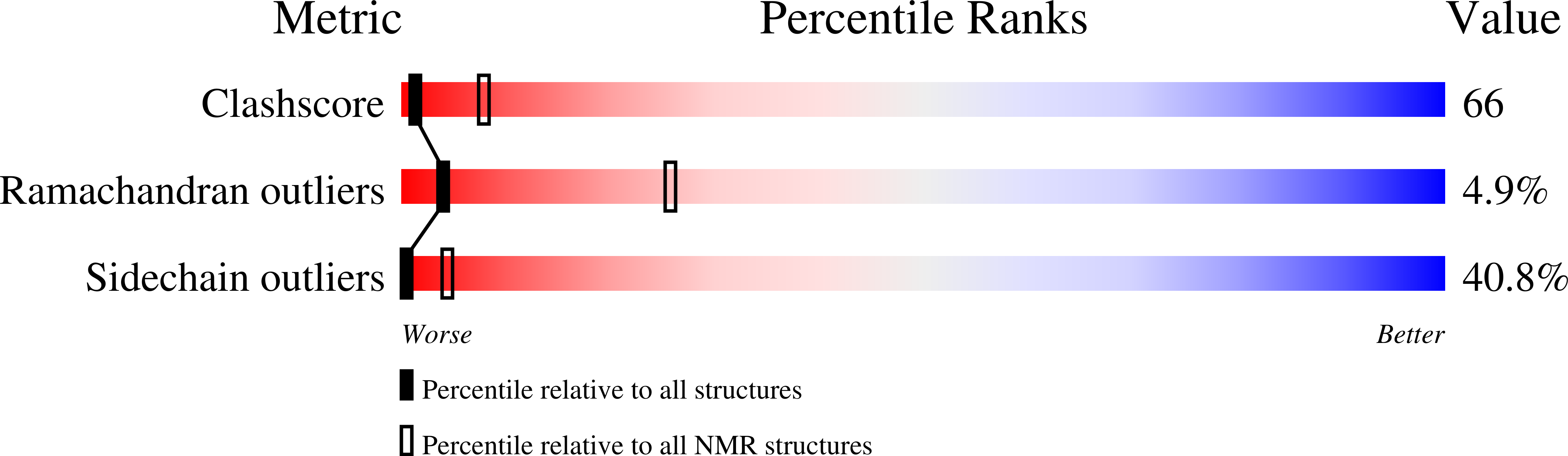Unmasking the Annexin I Interaction from the structure of apo-S100A11
Dempsey, A.C., Walsh, M.P., Shaw, G.S.(2003) Structure 11: 887-897
- PubMed: 12842051
- DOI: https://doi.org/10.1016/s0969-2126(03)00126-6
- Primary Citation of Related Structures:
1NSH - PubMed Abstract:
S100A11 is a homodimeric EF-hand calcium binding protein that undergoes a calcium-induced conformational change and interacts with the phospholipid binding protein annexin I to coordinate membrane association. In this work, the solution structure of apo-S100A11 has been determined by NMR spectroscopy to uncover the details of its calcium-induced structural change. Apo-S100A11 forms a tight globular structure having a near antiparallel orientation of helices III and IV in calcium binding site II. Further, helices I and IV, and I and I', form a more closed arrangement than observed in other apo-S100 proteins. This helix arrangement in apo-S100A11 partially buries residues in helices I (P3, E11, A15), III (V55, R58, M59), and IV (A86, C87, S90) and the linker (A45, F46), which are required for interaction with annexin I in the calcium-bound state. In apo-S100A11, this results in a "masked" binding surface that prevents annexin I binding but is uncovered upon calcium binding.
Organizational Affiliation:
Department of Biochemistry, The University of Western Ontario, London, Ontario N6A 5C1, Canada.