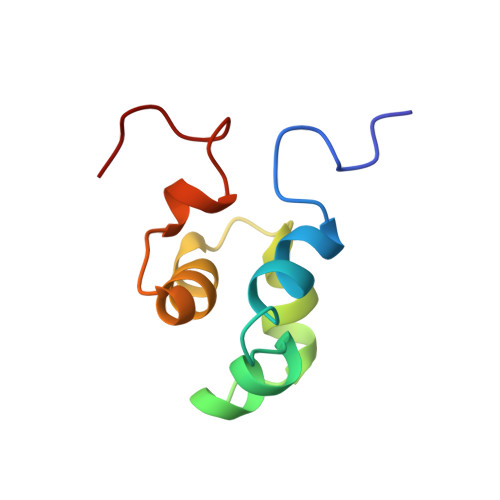
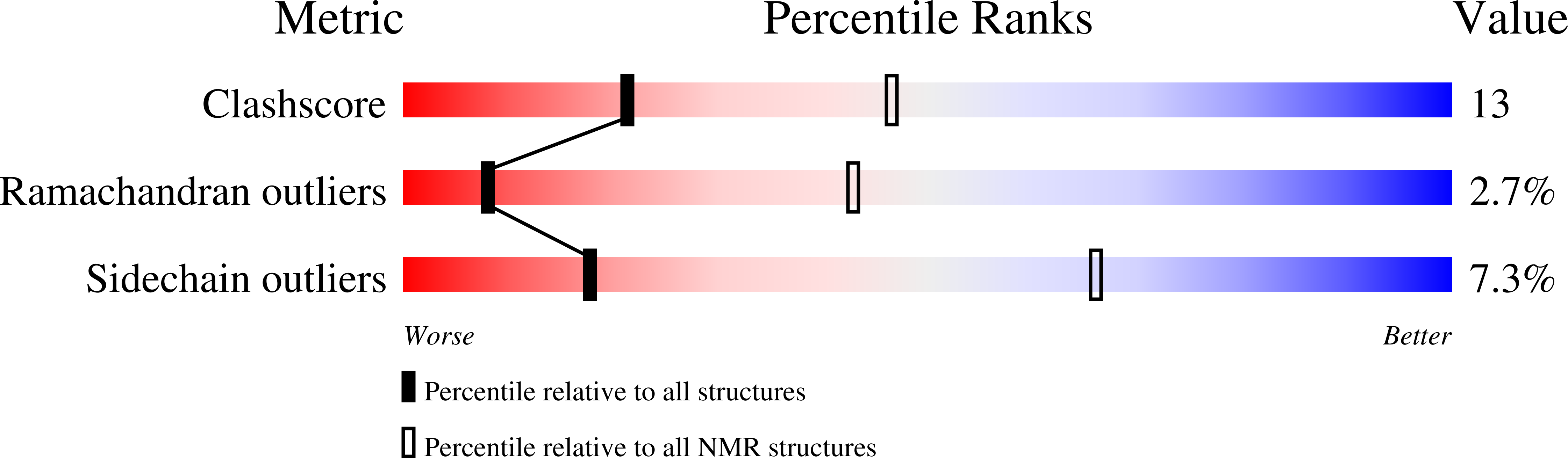The solution structure of the Mu Ner protein reveals a helix-turn-helix DNA recognition motif.
Strzelecka, T.E., Clore, G.M., Gronenborn, A.M.(1995) Structure 3: 1087-1095
- PubMed: 8590003
- DOI: https://doi.org/10.1016/s0969-2126(01)00244-1
- Primary Citation of Related Structures:
1NEQ, 1NER - PubMed Abstract:
The Mu Ner protein is a small (74 amino acids), basic, DNA-binding protein found in phage Mu. It belongs to a class of proteins, the cro and repressor proteins, that regulate the switch from the lysogenic to the lytic state of the phage life cycle. There is no significant sequence identity between Mu Ner and the cro proteins of other phages, despite their functional similarity. In addition, there is no significant sequence identity with any other DNA-binding proteins, with the exception of Ner from the related phage D108 and the Nlp protein of Escherichia coli. As the tertiary structures of Mu Ner and these two related proteins are unknown, it is clear that a three-dimensional (3D) structure of Mu Ner is essential in order to gain insight into its mode of DNA binding. The 3D solution structure of Mu Ner has been solved by 3D and 4D heteronuclear magnetic resonance spectroscopy. The structure consists of five alpha helices, two of which comprise a helix-turn-helix (HTH) motif. Analysis of line broadening and disappearance of crosspeaks in a 1H-15N correlation spectrum of the Mu Ner-DNA complex suggests that residues in these two helices are most likely to be in contact with the DNA. Like the functionally analogous cro proteins from phages lambda and 434, the Mu Ner protein possesses a HTH DNA recognition motif. The Ner protein from phage D108 and the Nlp protein from E. coli are likely to have very similar tertiary structures due to high amino-acid-sequence identity with Mu Ner.
Organizational Affiliation:
Laboratory of Chemical Physics, National Institutes of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892-0520, USA.