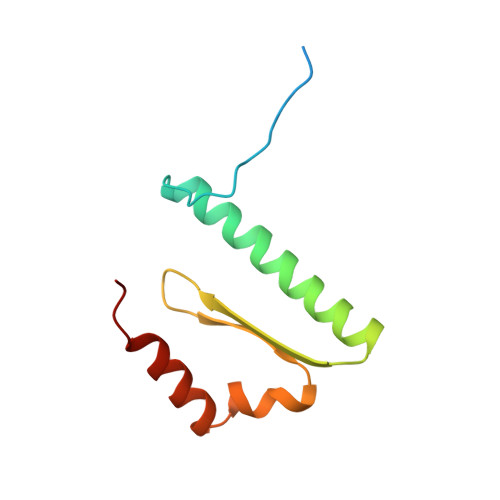
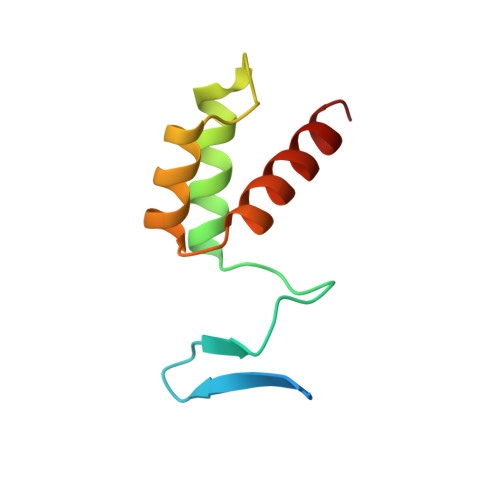
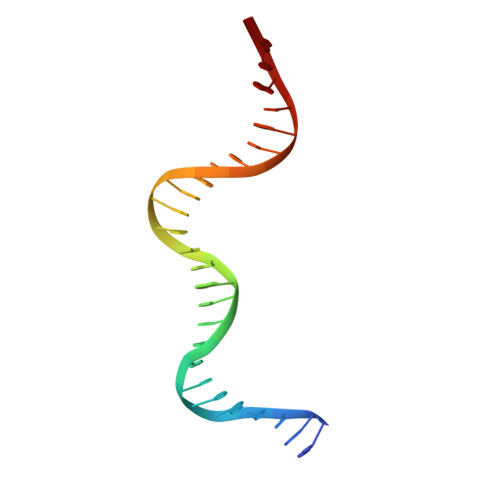
Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex.
Tan, S., Richmond, T.J.(1998) Nature 391: 660-666
- PubMed: 9490409
- DOI: https://doi.org/10.1038/35563
- Primary Citation of Related Structures:
1MNM - PubMed Abstract:
The structure of a complex containing the homeodomain repressor protein MATalpha2 and the MADS-box transcription factor MCM1 bound to DNA has been determined by X-ray crystallography at 2.25 A resolution. It reveals the protein-protein interactions responsible for cooperative binding of MATalpha2 and MCM1 to DNA. The otherwise flexible amino-terminal extension of the MATalpha2 homeodomain forms a beta-hairpin that grips the MCM1 surface through parallel beta-strand hydrogen bonds and close-packed, predominantly hydrophobic, side chains. DNA bending induced by MCM1 brings the two proteins closer together, facilitating their interaction. An unusual feature of the complex is that an eight-amino-acid sequence adopts an alpha-helical conformation in one of two copies of the MATalpha2 monomer and a beta-strand conformation in the other. This 'chameleon' sequence of MATalpha2 may be important for recognizing natural operator sites.
Organizational Affiliation:
ETH-Zurich, Institut für Molekularbiologie und Biophysik, Switzerland.