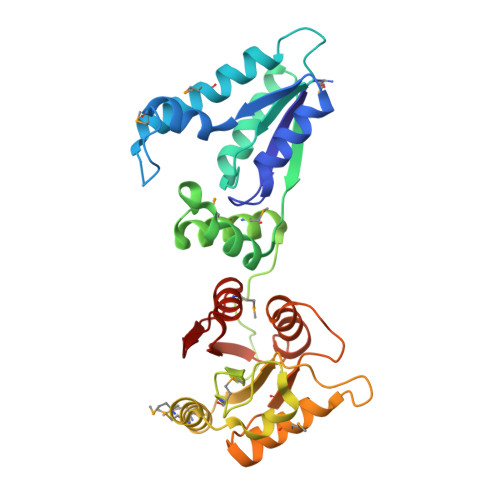
Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase.
Galburt, E.A., Pelletier, J., Wilson, G., Stoddard, B.L.(2002) Structure 10: 1249-1260
- PubMed: 12220496
- DOI: https://doi.org/10.1016/s0969-2126(02)00835-3
- Primary Citation of Related Structures:
1LTQ - PubMed Abstract:
T4 phage polynucleotide kinase (PNK) was identified over 35 years ago and has become a staple reagent for molecular biologists. The enzyme displays 5'-hydroxyl kinase, 3'-phosphatase, and 2',3'-cyclic phosphodiesterase activities against a wide range of substrates. These activities modify the ends of nicked tRNA generated by a bacterial response to infection and facilitate repair by T4 RNA ligase. DNA repair enzymes that share conserved motifs with PNK have been identified in eukaryotes. PNK contains two functionally distinct structural domains and forms a homotetramer. The C-terminal phosphatase domain is homologous to the L-2-haloacid dehalogenase family and the N-terminal kinase domain is homologous to adenylate kinase. The active sites have been characterized through structural homology analyses and visualization of bound substrate.
Organizational Affiliation:
Fred Hutchinson Cancer Research Center and The Graduate Program in Biomolecular Structure and Design, University of Washington, 1100 Fairview Avenue North, A3-023, Seattle, WA 98109, USA.