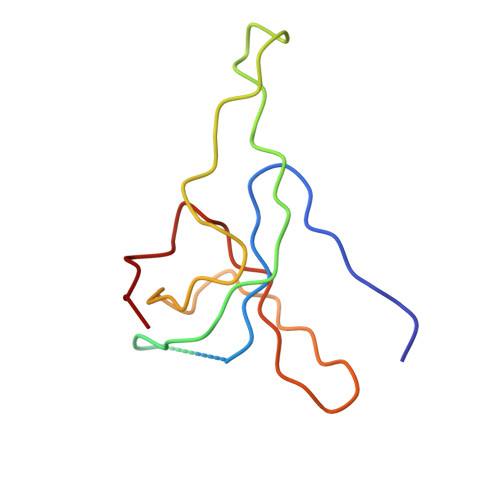
Structure of the heat shock protein chaperonin-10 of Mycobacterium leprae.
Mande, S.C., Mehra, V., Bloom, B.R., Hol, W.G.(1996) Science 271: 203-207
- PubMed: 8539620
- DOI: https://doi.org/10.1126/science.271.5246.203
- Primary Citation of Related Structures:
1LEP - PubMed Abstract:
Members of the chaperonin-10 (cpn10) protein family, also called heat shock protein 10 and in Escherichia coli GroES, play an important role in ensuring the proper folding of many proteins. The crystal structure of the Mycobacterium leprae cpn10 (Ml-cpn10) oligomer has been elucidated at a resolution of 3.5 angstroms. The architecture of the Ml-cpn10 heptamer resembles a dome with an oculus in its roof. The inner surface of the dome is hydrophilic and highly charged. A flexible region, known to interact with cpn60, extends from the lower rim of the dome. With the structure of a cpn10 heptamer now revealed and the structure of the E. coli GroEL previously known, models of cpn10:cpn60 and GroEL:GroES complexes are proposed.
Organizational Affiliation:
Department of Biological Structure, University of Washington, Seattle 98195, USA.