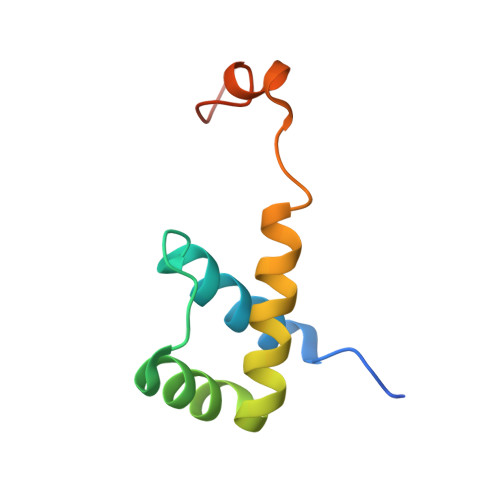
Structural and Thermodynamic Characterization of the DNA Binding Properties of a Triple Alanine Mutant of MATalpha2
Ke, A., Mathias, J.R., Vershon, A.K., Wolberger, C.(2002) Structure 10: 961-971
- PubMed: 12121651
- DOI: https://doi.org/10.1016/s0969-2126(02)00790-6
- Primary Citation of Related Structures:
1LE8 - PubMed Abstract:
Triply mutated MATalpha2 protein, alpha2-3A, in which all three major groove-contacting residues are mutated to alanine, is defective in binding DNA alone or in complex with Mcm1 yet binds with MATa1 with near wild-type affinity and specificity. To gain insight into this unexpected behavior, we determined the crystal structure of the a1/alpha2-3A/DNA complex. The structure shows that the triple mutation causes a collapse of the alpha2-3A/DNA interface that results in a reorganized set of alpha2-3A/DNA contacts, thereby enabling the mutant protein to recognize the wild-type DNA sequence. Isothermal titration calorimetry measurements reveal that a much more favorable entropic component stabilizes the a1/alpha2-3A/DNA complex than the alpha2-3A/DNA complex. The combined structural and thermodynamic studies provide an explanation of how partner proteins influence the sequence specificity of a DNA binding protein.
Organizational Affiliation:
Department of Biophysics and Biophysical Chemistry, John Hopkins University, 725 North Wolfe Street, Baltimore, MD 21205, USA.