A new family of plant transcription factors displays a novel ssDNA-binding surface.
Desveaux, D., Allard, J., Brisson, N., Sygusch, J.(2002) Nat Struct Biol 9: 512-517
- PubMed: 12080340
- DOI: https://doi.org/10.1038/nsb814
- Primary Citation of Related Structures:
1L3A - PubMed Abstract:
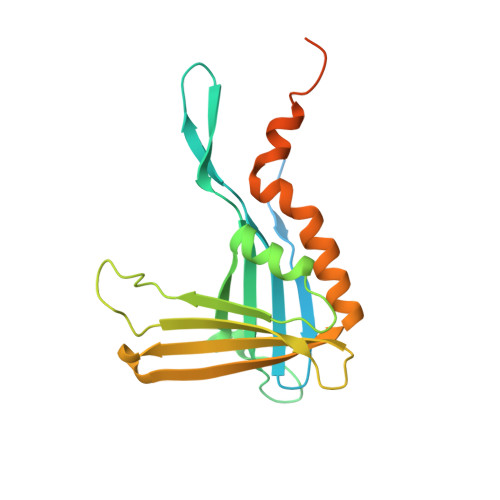
The crystal structure of p24, the single-stranded DNA (ssDNA) binding subunit of the plant defense transcription factor PBF-2, has been determined to 2.3 A resolution. p24 is representative of a novel family of ubiquitous plant-specific proteins that we refer to as the Whirly family because of their quaternary structure. PBF-2 is composed of four p24 molecules that interact through a helix-loop-helix motif. This interaction produces a central pore, with beta-strands radiating outwards, resulting in a whirligig appearance to the quaternary structure. The noncrystallographic C(4) symmetry arrangement of p24 subunits is novel for ssDNA binding proteins and may explain the binding specificity of PBF-2. This structural arrangement also supports the role of PBF-2 in binding melted promoter regions to modulate gene expression.
Organizational Affiliation:
Department of Biochemistry, Université de Montréal, CP 6128, Station Centre-Ville, Montréal, Québec, H3C 3J7, Canada.