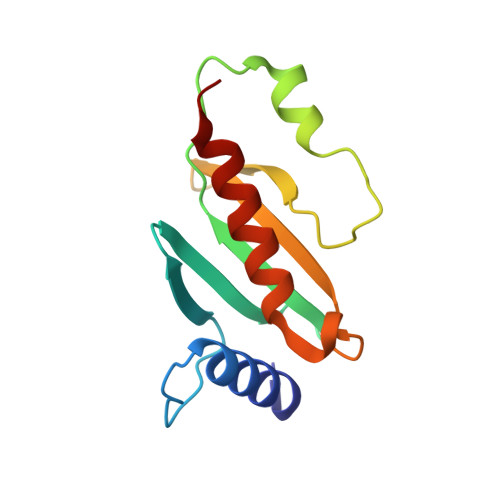
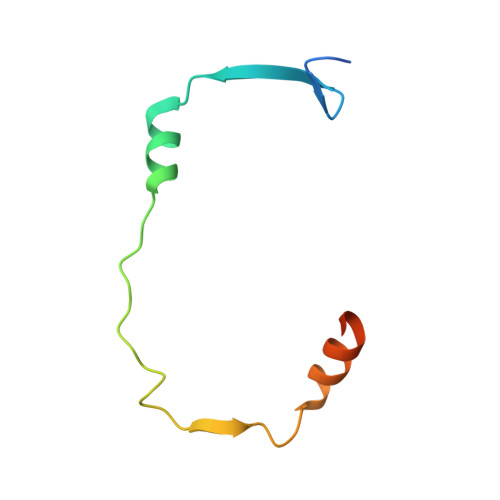
Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens.
Birtalan, S.C., Phillips, R.M., Ghosh, P.(2002) Mol Cell 9: 971-980
- PubMed: 12049734
- DOI: https://doi.org/10.1016/s1097-2765(02)00529-4
- Primary Citation of Related Structures:
1L2W - PubMed Abstract:
The type III secretion system (TTSS) of Gram-negative bacterial pathogens delivers effector proteins required for virulence directly into the cytosol of host cells. Delivery of many effectors depends on association with specific cognate chaperones in the bacterial cytosol. The mechanism of chaperone action is not understood. Here we present biochemical and crystallographic results on the Yersinia SycE-YopE chaperone-effector complex that contradict previous models of chaperone function and demonstrate that chaperone action is isolated to only a small portion of the effector. This, together with evidence for stereochemical conservation between chaperone-effector complexes, which are otherwise unrelated in sequence, indicates that these complexes function as general, three-dimensional TTSS secretion signals and may endow a temporal order to secretion.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA 92093, USA.