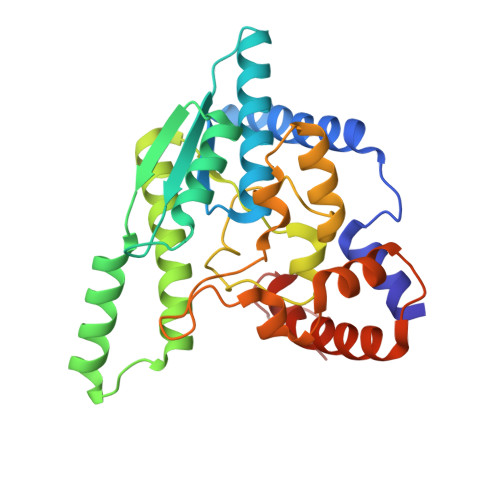
NH3-dependent NAD+ synthetase from Bacillus subtilis at 1 A resolution.
Symersky, J., Devedjiev, Y., Moore, K., Brouillette, C., DeLucas, L.(2002) Acta Crystallogr D Biol Crystallogr 58: 1138-1146
- PubMed: 12077433
- DOI: https://doi.org/10.1107/s0907444902006698
- Primary Citation of Related Structures:
1KQP - PubMed Abstract:
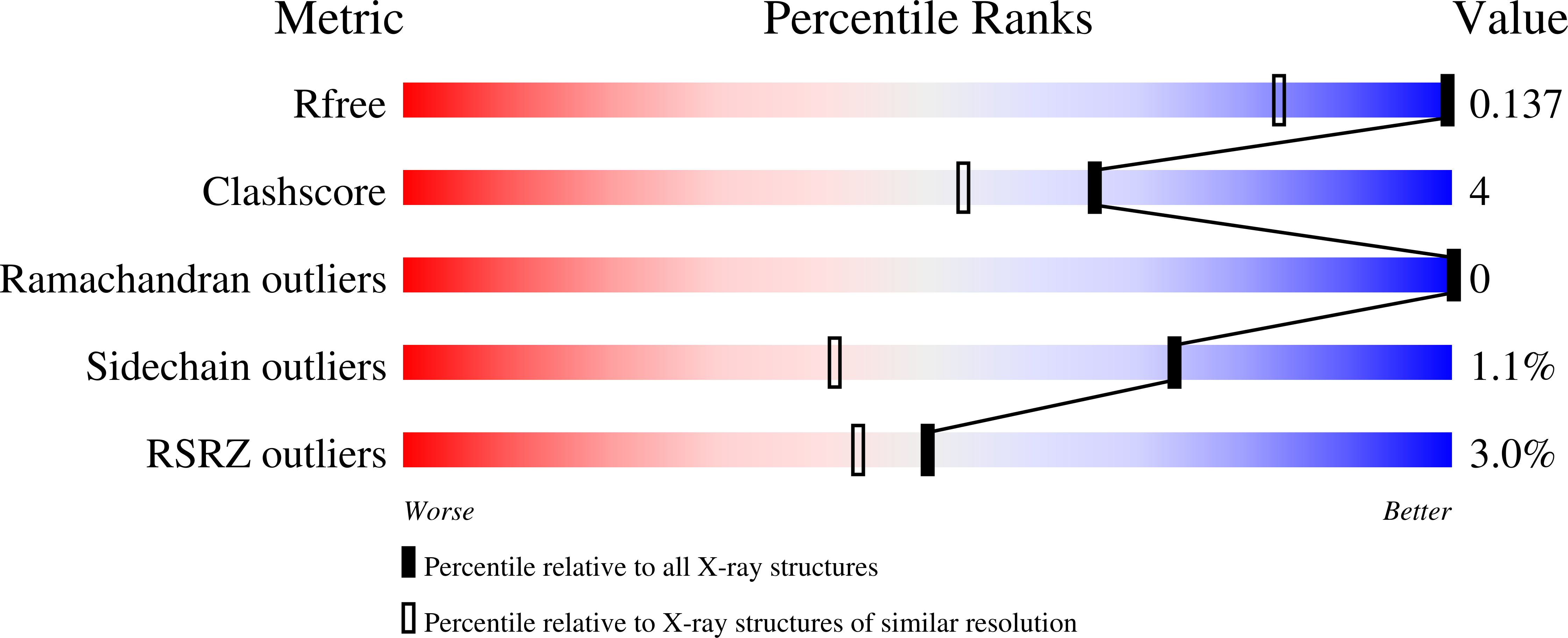
The final step of NAD+ biosynthesis includes an amide transfer to nicotinic acid adenine dinucleotide (NaAD) catalyzed by NAD+ synthetase. This enzyme was co-crystallized in microgravity with natural substrates NaAD and ATP at pH 8.5. The crystal was exposed to ammonium ions, synchrotron diffraction data were collected and the atomic model was refined anisotropically at 1 A resolution to R = 11.63%. Both binding sites are occupied by the NAD-adenylate intermediate, pyrophosphate and two magnesium ions. The atomic resolution of the structure allows better definition of non-planar peptide groups, reveals a low mean anisotropy of protein and substrate atoms and indicates the H-atom positions of the phosphoester group of the reaction intermediate. The phosphoester group is protonated at the carbonyl O atom O7N, suggesting a carbenium-ion structure stabilized by interactions with two solvent sites presumably occupied by ammonia and a water molecule. A mechanism is proposed for the second catalytic step, which includes a nucleophilic attack by the ammonia molecule on the intermediate.
Organizational Affiliation:
Center for Biophysical Sciences and Engineering, University of Alabama at Birmingham, 35294, USA.