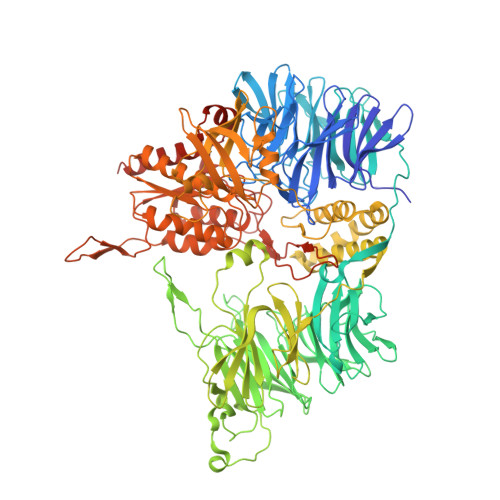
Crystal structure of the tricorn protease reveals a protein disassembly line.
Brandstetter, H., Kim, J.S., Groll, M., Huber, R.(2001) Nature 414: 466-470
- PubMed: 11719810
- DOI: https://doi.org/10.1038/35106609
- Primary Citation of Related Structures:
1K32 - PubMed Abstract:
The degradation of cytosolic proteins is carried out predominantly by the proteasome, which generates peptides of 7-9 amino acids long. These products need further processing. Recently, a proteolytic system was identified in the model organism Thermoplasma acidophilum that performs this processing. The hexameric core protein of this modular system, referred to as tricorn protease, is a 720K protease that is able to assemble further into a giant icosahedral capsid, as determined by electron microscopy. Here, we present the crystal structure of the tricorn protease at 2.0 A resolution. The structure reveals a complex mosaic protein whereby five domains combine to form one of six subunits, which further assemble to form the 3-2-symmetric core protein. The structure shows how the individual domains coordinate the specific steps of substrate processing, including channelling of the substrate to, and the product from, the catalytic site. Moreover, the structure shows how accessory protein components might contribute to an even more complex protein machinery that efficiently collects the tricorn-released products.