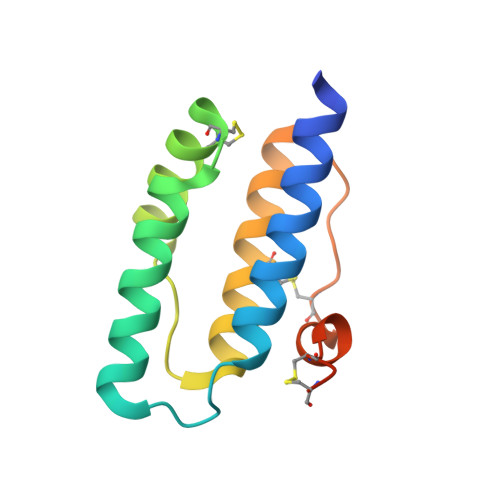
A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p.
Gross, E., Sevier, C.S., Vala, A., Kaiser, C.A., Fass, D.(2002) Nat Struct Biol 9: 61-67
- PubMed: 11740506
- DOI: https://doi.org/10.1038/nsb740
- Primary Citation of Related Structures:
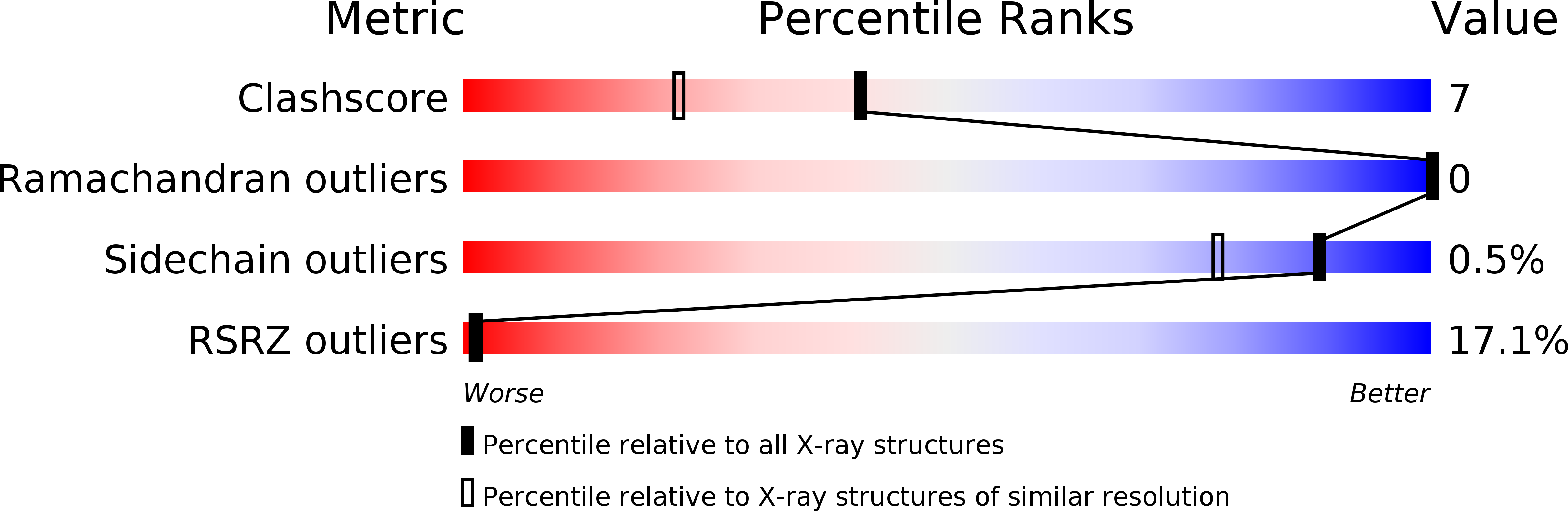
1JR8, 1JRA - PubMed Abstract:
Erv2p is an FAD-dependent sulfhydryl oxidase that can promote disulfide bond formation during protein biosynthesis in the yeast endoplasmic reticulum. The structure of Erv2p, determined by X-ray crystallography to 1.5 A resolution, reveals a helix-rich dimer with no global resemblance to other known FAD-binding proteins or thiol oxidoreductases. Two pairs of cysteine residues are required for Erv2p activity. The first (Cys-Gly-Glu-Cys) is adjacent to the isoalloxazine ring of the FAD. The second (Cys-Gly-Cys) is part of a flexible C-terminal segment that can swing into the vicinity of the first cysteine pair in the opposite subunit of the dimer and may shuttle electrons between substrate protein dithiols and the FAD-proximal disulfide.
Organizational Affiliation:
Department of Structural Biology, Weizmann Institute of Science, Rehovot 76100, Israel.