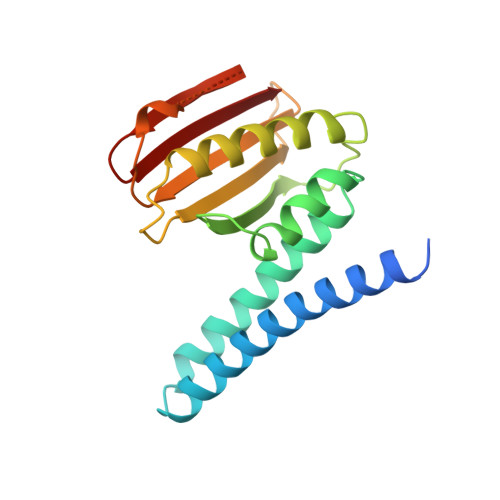
Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase.
Varughese, K.I., Madhusudan, Zhou, X.Z., Whiteley, J.M., Hoch, J.A.(1998) Mol Cell 2: 485-493
- PubMed: 9809070
- DOI: https://doi.org/10.1016/s1097-2765(00)80148-3
- Primary Citation of Related Structures:
1IXM - PubMed Abstract:
A basis for understanding specificity of molecular recognition between phosphorelay proteins has been deduced from the 2.6 A structure of the Spo0B phosphotransferase of the phosphorelay regulating sporulation initiation. Spo0B consists of two domains: an N-terminal alpha-helical hairpin domain and a C-terminal alpha/beta domain. Two subunits of Spo0B dimerize by a parallel association of helical hairpins to form a novel four-helix bundle from which the active histidine protrudes. Docking studies show that both the monomers interact with a Spo0F molecule at the region surrounding the active site aspartate to position it for phosphotransfer. It is apparent that different surfaces of response regulators may be involved in recognition of the protein partners to which they are paired.
Organizational Affiliation:
Department of Molecular and Experimental Medicine, Scripps Research Institute, La Jolla, California 92037, USA. kiv@scripps.edu