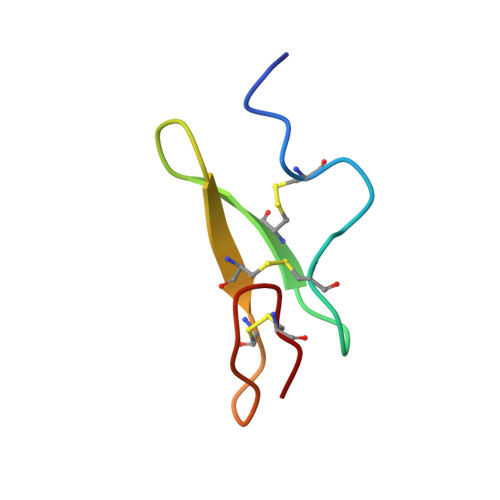
The three-dimensional structure of the first EGF-like module of human factor IX: comparison with EGF and TGF-alpha.
Baron, M., Norman, D.G., Harvey, T.S., Handford, P.A., Mayhew, M., Tse, A.G., Brownlee, G.G., Campbell, I.D.(1992) Protein Sci 1: 81-90
- PubMed: 1304885
- DOI: https://doi.org/10.1002/pro.5560010109
- Primary Citation of Related Structures:
1IXA - PubMed Abstract:
The three-dimensional structure of the first epidermal growth factor (EGF)-like module from human factor IX has been determined in solution using two-dimensional nuclear magnetic resonance (in the absence of calcium and at pH 4.5). The structure was found to resemble closely that of EGF and the homologous transforming growth factor-alpha (TGF-alpha). Residues 60-65 form an antiparallel beta-sheet with residues 68-73. In the C-terminal subdomain a type II beta-turn is found between residues 74 and 77 and a five-residue turn is found between residues 79 and 83. Glu 78 and Leu 84 pair in an antiparallel beta-sheet conformation. In the N-terminal region a loop is found between residues 50 and 55 such that the side chains of both are positioned above the face of the beta-sheet. Residues 56-60 form a turn that leads into the first strand of the beta-sheet. Whereas the global fold closely resembles that of EGF, the N-terminal residues of the module (46-49) do not form a beta-strand but are ill-defined in the structure, probably due to the local flexibility of this region. The structure is discussed with reference to recent site-directed mutagenesis data, which have identified certain conserved residues as ligands for calcium.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, UK.