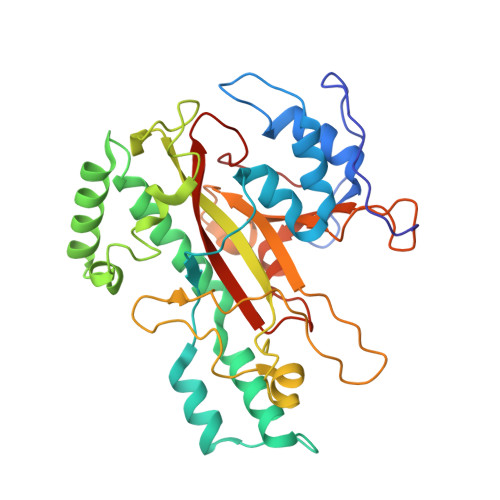
Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense
Kashiwagi, T., Yokoyama, K., Ishikawa, K., Ono, K., Ejima, D., Matsui, H., Suzuki, E.(2002) J Biol Chem 277: 44252-44260
- PubMed: 12221081
- DOI: https://doi.org/10.1074/jbc.M203933200
- Primary Citation of Related Structures:
1IU4 - PubMed Abstract:
The crystal structure of a microbial transglutaminase from Streptoverticillium mobaraense has been determined at 2.4 A resolution. The protein folds into a plate-like shape, and has one deep cleft at the edge of the molecule. Its overall structure is completely different from that of the factor XIII-like transglutaminase, which possesses a cysteine protease-like catalytic triad. The catalytic residue, Cys(64), exists at the bottom of the cleft. Asp(255) resides at the position nearest to Cys(64) and is also adjacent to His(274). Interestingly, Cys(64), Asp(255), and His(274) superimpose well on the catalytic triad "Cys-His-Asp" of the factor XIII-like transglutaminase, in this order. The secondary structure frameworks around these residues are also similar to each other. These results imply that both transglutaminases are related by convergent evolution; however, the microbial transglutaminase has developed a novel catalytic mechanism specialized for the cross-linking reaction. The structure accounts well for the catalytic mechanism, in which Asp(255) is considered to be enzymatically essential, as well as for the causes of the higher reaction rate, the broader substrate specificity, and the lower deamidation activity of this enzyme.
Organizational Affiliation:
Central Research Laboratories, Ajinomoto Company Inc., 1-1 Suzuki-cho, Kawasaki-ku, Kanagawa 210-8681, Japan.