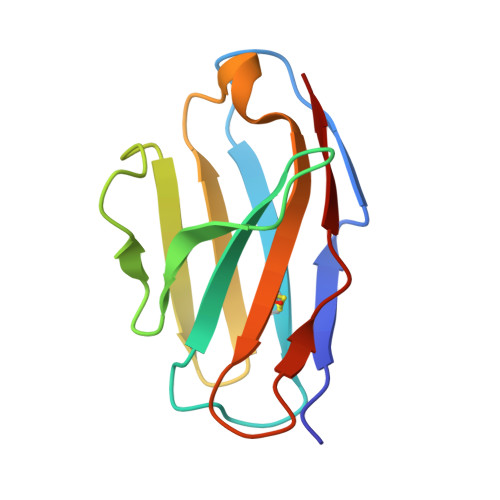
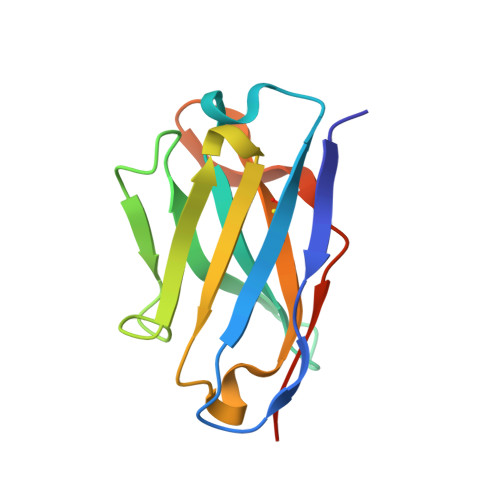
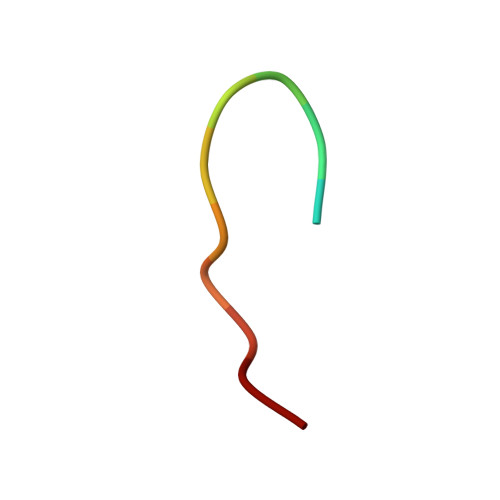
Antibody recognition of a conformational epitope in a peptide antigen: Fv-peptide complex of an antibody fragment specific for the mutant EGF receptor, EGFRvIII.
Landry, R.C., Klimowicz, A.C., Lavictoire, S.J., Borisova, S., Kottachchi, D.T., Lorimer, I.A., Evans, S.V.(2001) J Mol Biol 308: 883-893
- PubMed: 11352579
- DOI: https://doi.org/10.1006/jmbi.2001.4628
- Primary Citation of Related Structures:
1I8I, 1I8K - PubMed Abstract:
Epitope mapping studies and the determination of the structure to 1.8 A resolution have been carried out for the antigen-binding fragment MR1 in complex with peptide antigen. MR1 is specific for the novel fusion junction of the mutant epidermal growth factor receptor EGFRvIII and has been reported to have a high degree of specificity for the mutant EGFRvIII over the wild-type EGF receptor. The structure of the complex shows that the peptide antigen residue side-chains found by epitope mapping studies to be critical for recognition are accommodated in pockets on the surface of the Fv. However, the most distinctive portion of the peptide antigen, the novel fusion glycine residue, makes no contact to the Fv and does not contribute directly to the epitope. The specificity of MR1 lies in the ability of this glycine residue to assume the restricted conformation needed to form a type II' beta-hairpin turn more easily, and demonstrates that a peptide antigen can be used to generate a conformational epitope.
Organizational Affiliation:
Department of Biochemistry Microbiology and Immunology, University of Ottawa, 451 Smyth, Ottawa, K1H 8M5, Canada.