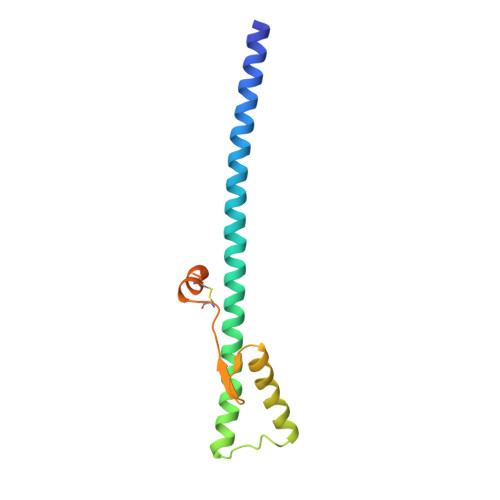
Structure of influenza haemagglutinin at the pH of membrane fusion.
Bullough, P.A., Hughson, F.M., Skehel, J.J., Wiley, D.C.(1994) Nature 371: 37-43
- PubMed: 8072525
- DOI: https://doi.org/10.1038/371037a0
- Primary Citation of Related Structures:
1HTM - PubMed Abstract:
Low pH induces a conformational change in the influenza virus haemagglutinin, which then mediates fusion of the viral and host cell membranes. The three-dimensional structure of a fragment of the haemagglutinin in this conformation reveals a major refolding of the secondary and tertiary structure of the molecule. The apolar fusion peptide moves at least 100 A to one tip of the molecule. At the other end a helical segment unfolds, a subdomain relocates reversing the chain direction, and part of the structure becomes disordered.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Harvard University, Cambridge, Massachusetts 02138.