The bacterial periplasmic histidine-binding protein. structure/function analysis of the ligand-binding site and comparison with related proteins.
Oh, B.H., Kang, C.H., De Bondt, H., Kim, S.H., Nikaido, K., Joshi, A.K., Ames, G.F.(1994) J Biol Chem 269: 4135-4143
- PubMed: 8307974
- Primary Citation of Related Structures:
1HPB - PubMed Abstract:
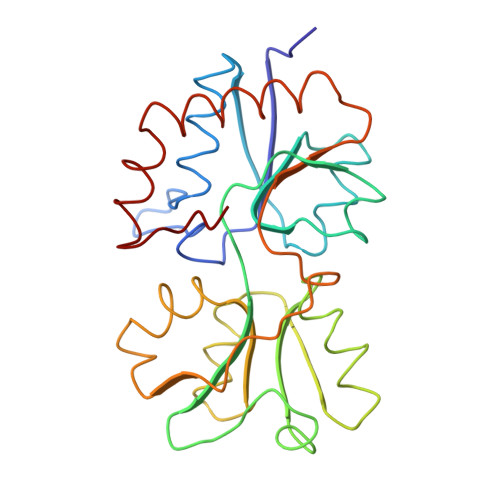
Bacterial periplasmic binding proteins are initial receptors in the process of active transport across cell membranes and/or chemotaxis. Among them, the histidine-binding protein (HisJ) has been extensively studied from the biochemical, physiological, and genetic points of view. The three-dimensional crystal structure of the histidine-binding protein complexed with histidine has been determined at 2.5-A resolution by the molecular replacement method using a probe structure the previously solved lysine-liganded structure of the lysine-, arginine-, ornithine-binding protein (LAO), which shares 70% sequence identity with HisJ. The structure is bi-lobate; the two lobes, one bigger than the other, are connected by two short strands and are in contact with each other (closed) enclosing the histidine. Charged, polar, and non-polar side chains, as well as the peptide backbone, are involved in tight binding of the histidine. The bound histidine is involved in eight direct hydrogen bonds, six with the bigger lobe and two with the smaller lobe, in one potential water-mediated hydrogen bond with the bigger lobe, as well as in ionic interactions. The HisJ residues surrounding the ligand are the same as the LAO residues interacting with lysine, except for residue 52 which is leucine in HisJ and phenylalanine in LAO. The Leu-52 in HisJ makes a hydrophobic interaction with the imidazole ring of histidine. Of seven mutations affecting the ligand-binding site, five are located in the ligand-binding site, one in a connecting strand, and one at the domains interface. Based on comparisons among related binding proteins, the specific interactions between the ligands and the respective binding protein residues are predicted for the glutamine-binding protein and the opines-binding protein.
Organizational Affiliation:
Department of Chemistry, University of California, Berkeley 94720.