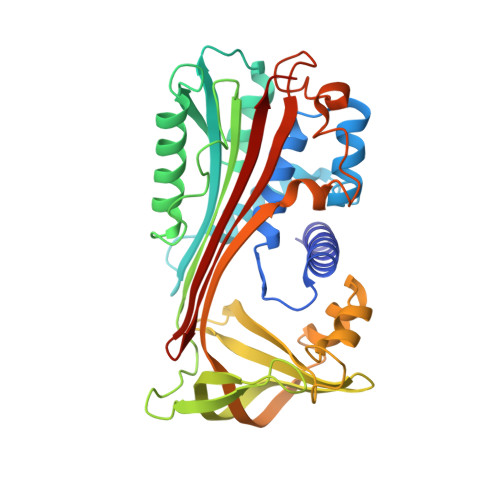
Crystal structure of cleaved equine leucocyte elastase inhibitor determined at 1.95 A resolution.
Baumann, U., Bode, W., Huber, R., Travis, J., Potempa, J.(1992) J Mol Biol 226: 1207-1218
- PubMed: 1518052
- DOI: https://doi.org/10.1016/0022-2836(92)91062-t
- Primary Citation of Related Structures:
1HLE - PubMed Abstract:
The crystal structure of active-site cleaved equine leucocyte elastase inhibitor, a member of the serpin superfamily, has been solved and refined to a crystallographic R-factor of 17.6% at 1.95 A resolution. Despite being an intracellular inhibitor with rather low sequence homology of 30% to human alpha 1-antichymotrypsin and alpha 1-proteinase inhibitor, the three-dimensional structures are very similar, with deviations only at the sites of insertions and few mobile secondary structure elements. The better resolution in comparison with the structures of other cleaved serpins allows a more precise description of the so-called R-state of the serpins.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Martinsried bei München, Germany.