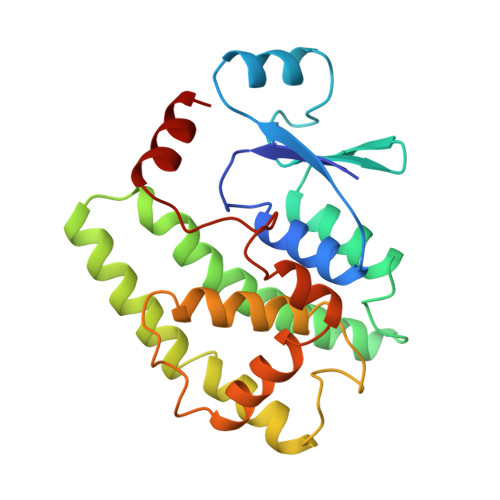
Crystal structure of a murine alpha-class glutathione S-transferase involved in cellular defense against oxidative stress.
Krengel, U., Schroter, K.H., Hoier, H., Arkema, A., Kalk, K.H., Zimniak, P., Dijkstra, B.W.(1998) FEBS Lett 422: 285-290
- PubMed: 9498801
- DOI: https://doi.org/10.1016/s0014-5793(98)00026-x
- Primary Citation of Related Structures:
1GUK - PubMed Abstract:
Glutathione S-transferases (GSTs) are ubiquitous multifunctional enzymes which play a key role in cellular detoxification. The enzymes protect the cells against toxicants by conjugating them to glutathione. Recently, a novel subgroup of alpha-class GSTs has been identified with altered substrate specificity which is particularly important for cellular defense against oxidative stress. Here, we report the crystal structure of murine GSTA4-4, which is the first structure of a prototypical member of this subgroup. The structure was solved by molecular replacement and refined to 2.9 A resolution. It resembles the structure of other members of the GST superfamily, but reveals a distinct substrate binding site.
Organizational Affiliation:
BIOSON Research Institute, Department of Chemistry, University of Groningen, The Netherlands.