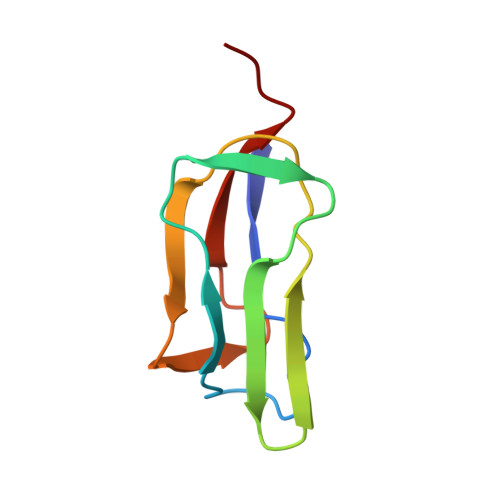
Solution Structure of the Lipoyl Domain of the Chimeric Dihydrolipoyl Dehydrogenase P64K from Neisseria Meningitidis
Tozawa, K., Broadhurst, R.W., Raine, A.R., Fuller, C., Alvarez, A., Guillen, G., Padron, G., Perham, R.N.(2001) Eur J Biochem 268: 4908
- PubMed: 11559360
- DOI: https://doi.org/10.1046/j.0014-2956.2001.02422.x
- Primary Citation of Related Structures:
1GJX - PubMed Abstract:
The antigenic P64K protein from the pathogenic bacterium Neisseria meningitidis is found in the outer membrane of the cell, and consists of two parts: an 81-residue N-terminal region and a 482-residue C-terminal region. The amino-acid sequence of the N-terminal region is homologous with the lipoyl domains of the dihydrolipoyl acyltransferase (E2) components, and that of the C-terminal region with the dihydrolipoyl dehydrogenase (E3) components, of 2-oxo acid dehydrogenase multienzyme complexes. The two parts are separated by a long linker region, similar to the linker regions in the E2 chains of 2-oxo acid dehydrogenase complexes, and it is likely this region is conformationally flexible. A subgene encoding the P64K lipoyl domain was created and over-expressed in Escherichia coli. The product was capable of post-translational modification by the lipoate protein ligase but not aberrant modification by the biotin protein ligase of E. coli. The solution structure of the apo-domain was determined by means of heteronuclear NMR spectroscopy and found to be a flattened beta barrel composed of two four-stranded antiparallel beta sheets. The lysine residue that becomes lipoylated is in an exposed beta turn that, from a [1H]-15N heteronuclear Overhauser effect experiment, appears to enjoy substantial local motion. This structure of a lipoyl domain derived from a dihydrolipoyl dehydrogenase resembles that of lipoyl domains normally found as part of the dihydrolipoyl acyltransferase component of 2-oxo acid dehydrogenase complexes and will assist in furthering the understanding of its function in a multienzyme complex and in the membrane-bound P64K protein itself.
Organizational Affiliation:
Cambridge Centre for Molecular Recognition, Department of Biochemistry, University of Cambridge, UK.