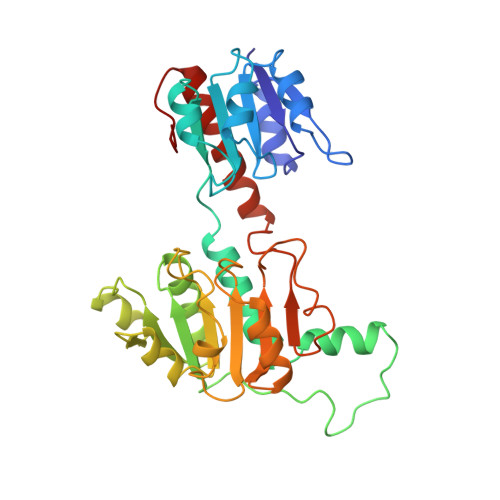
Crystal structure of a NAD-dependent D-glycerate dehydrogenase at 2.4 A resolution.
Goldberg, J.D., Yoshida, T., Brick, P.(1994) J Mol Biol 236: 1123-1140
- PubMed: 8120891
- DOI: https://doi.org/10.1016/0022-2836(94)90016-7
- Primary Citation of Related Structures:
1GDH - PubMed Abstract:
D-Glycerate dehydrogenase (GDH) catalyzes the NADH-linked reduction of hydroxypyruvate to D-glycerate. GDH is a member of a family of NAD-dependent dehydrogenases that is characterized by a specificity for the D-isomer of the hydroxyacid substrate. The crystal structure of the apoenzyme form of GDH from Hyphomicrobium methylovorum has been determined by the method of isomorphous replacement and refined at 2.4 A resolution using a restrained least-squares method. The crystallographic R-factor is 19.4% for all 24,553 measured reflections between 10.0 and 2.4 A resolution. The GDH molecule is a symmetrical dimer composed of subunits of molecular mass 38,000, and shares significant structural homology with another NAD-dependent enzyme, formate dehydrogenase. The GDH subunit consists of two structurally similar domains that are approximately related to each other by 2-fold symmetry. The domains are separated by a deep cleft that forms the putative NAD and substrate binding sites. One of the domains has been identified as the NAD-binding domain based on its close structural similarity to the NAD-binding domains of other NAD-dependent dehydrogenases. The topology of the second domain is different from that found in the various catalytic domains of other dehydrogenases. A model of a ternary complex of GDH has been built in which putative catalytic residues are identified based on sequence homology between the D-isomer specific dehydrogenases. A structural comparison between GDH and L-lactate dehydrogenase indicates a convergence of active site residues and geometries for these two enzymes. The reactions catalyzed are chemically equivalent but of opposing stereospecificity. A hypothesis is presented to explain how the two enzymes may exploit the same coenzyme stereochemistry and a similar spatial arrangement of catalytic residues to carry out reactions that proceed to opposite enantiomers.
Organizational Affiliation:
Blackett Laboratory, Imperial College, London, England.