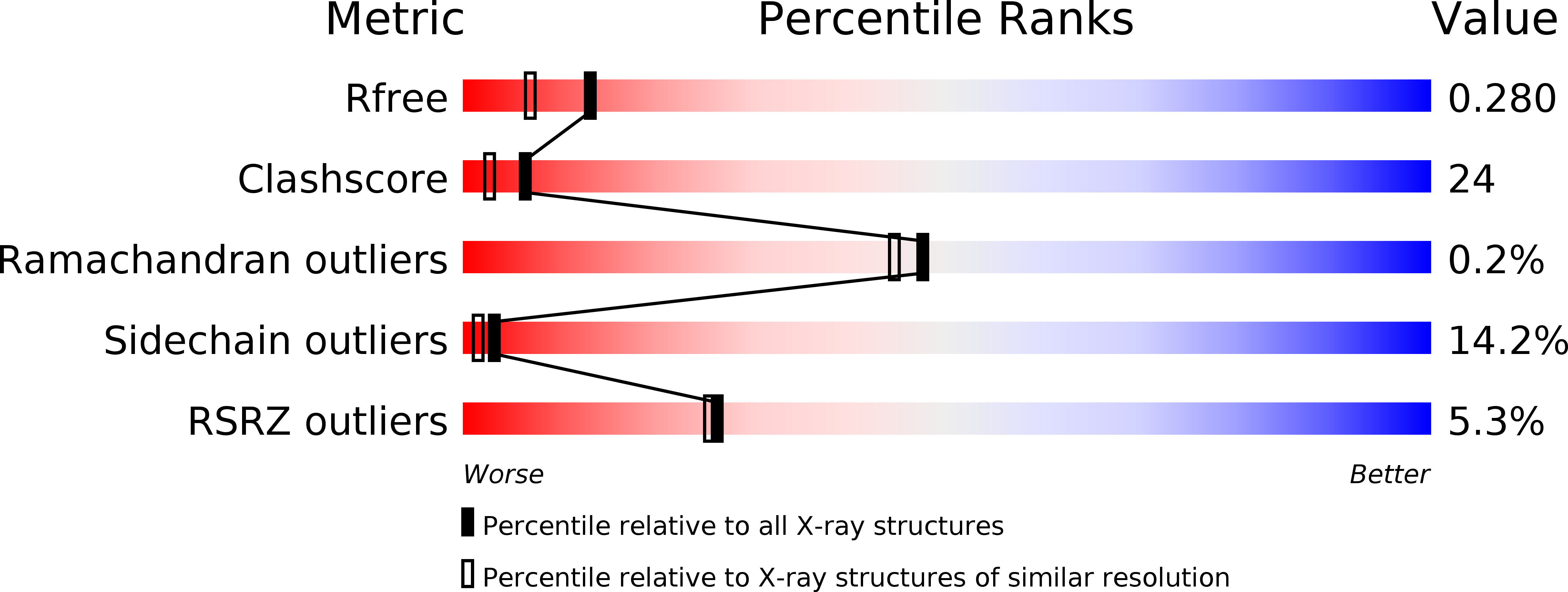
The crystal structure of the ttCsaA protein: an export-related chaperone from Thermus thermophilus.
Kawaguchi, S., Muller, J., Linde, D., Kuramitsu, S., Shibata, T., Inoue, Y., Vassylyev, D.G., Yokoyama, S.(2001) EMBO J 20: 562-569
- PubMed: 11157762
- DOI: https://doi.org/10.1093/emboj/20.3.562
- Primary Citation of Related Structures:
1GD7 - PubMed Abstract:
The CsaA protein was first characterized in Bacillus subtilis as a molecular chaperone with export-related activities. Here we report the 2.0 Angstrom-resolution crystal structure of the Thermus thermophilus CsaA protein, designated ttCsaA. Atomic structure and experiments in solution revealed a homodimer as the functional unit. The structure of the ttCsaA monomer is reminiscent of the well known oligonucleotide-binding fold, with the addition of extensions at the N- and C-termini that form an extensive dimer interface. The two identical, large, hydrophobic cavities on the protein surface are likely to constitute the substrate binding sites. The CsaA proteins share essential sequence similarity with the tRNA-binding protein Trbp111. Structure-based sequence analysis suggests a close structural resemblance between these proteins, which may extend to the architecture of the binding sites at the atomic level. These results raise the intriguing possibility that CsaA proteins possess a second, tRNA-binding activity in addition to their export-related function.
Organizational Affiliation:
RIKEN Harima Institute at SPring-8, 1-1-1 Kouto, Mikazuki-cho, Sayo-Gun, Hyogo 679-5148, Japan.