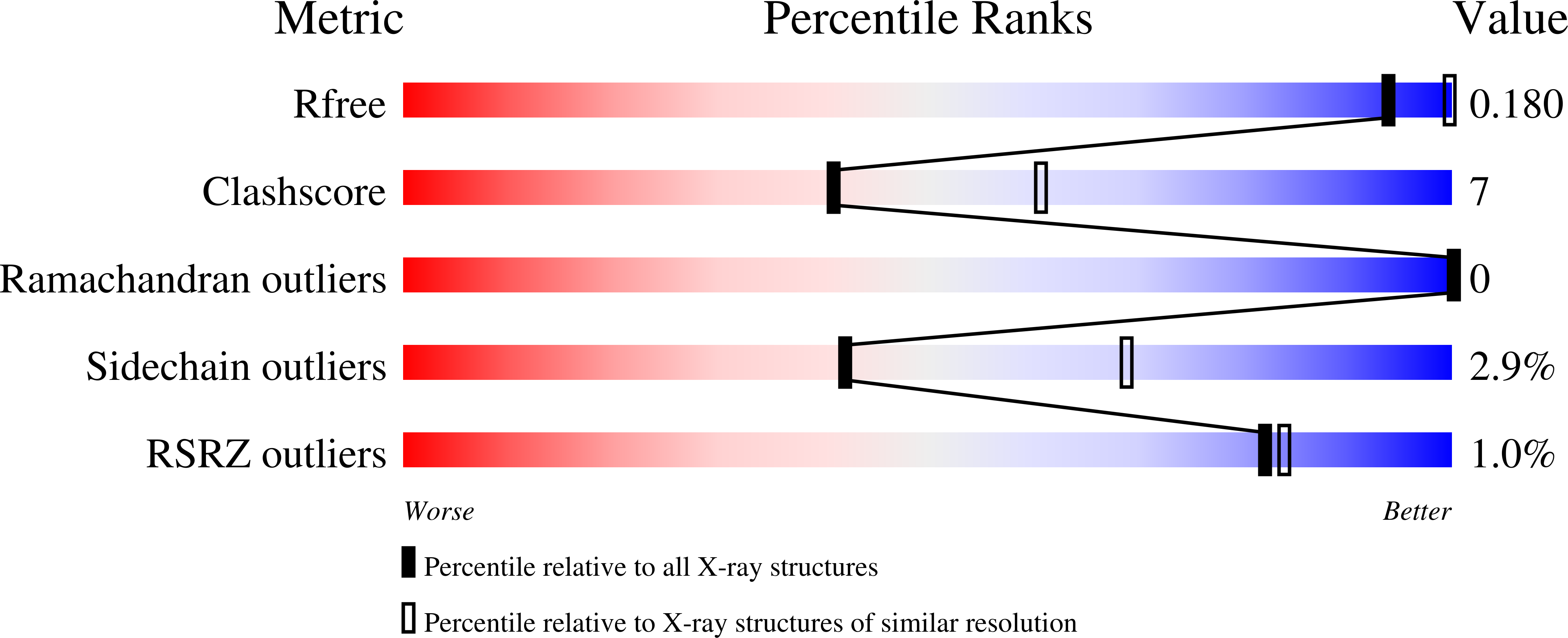
Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases.
Buss, K.A., Cooper, D.R., Ingram-Smith, C., Ferry, J.G., Sanders, D.A., Hasson, M.S.(2001) J Bacteriol 183: 680-686
- PubMed: 11133963
- DOI: https://doi.org/10.1128/JB.183.2.680-686.2001
- Primary Citation of Related Structures:
1G99 - PubMed Abstract:
Acetate kinase, an enzyme widely distributed in the Bacteria and Archaea domains, catalyzes the phosphorylation of acetate. We have determined the three-dimensional structure of Methanosarcina thermophila acetate kinase bound to ADP through crystallography. As we previously predicted, acetate kinase contains a core fold that is topologically identical to that of the ADP-binding domains of glycerol kinase, hexokinase, the 70-kDa heat shock cognate (Hsc70), and actin. Numerous charged active-site residues are conserved within acetate kinases, but few are conserved within the phosphotransferase superfamily. The identity of the points of insertion of polypeptide segments into the core fold of the superfamily members indicates that the insertions existed in the common ancestor of the phosphotransferases. Another remarkable shared feature is the unusual, epsilon conformation of the residue that directly precedes a conserved glycine residue (Gly-331 in acetate kinase) that binds the alpha-phosphate of ADP. Structural, biochemical, and geochemical considerations indicate that an acetate kinase may be the ancestral enzyme of the ASKHA (acetate and sugar kinases/Hsc70/actin) superfamily of phosphotransferases.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907, USA.