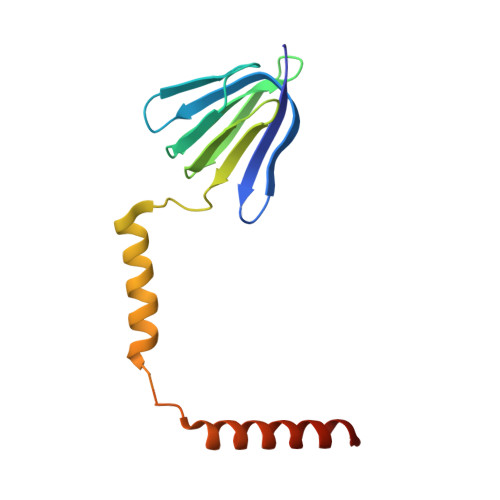
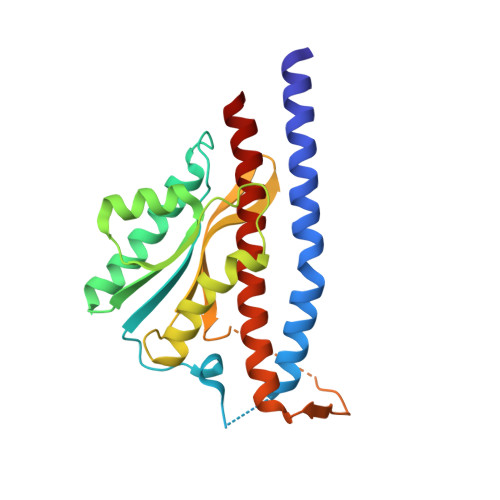
Structure of the gamma-epsilon complex of ATP synthase.
Rodgers, A.J., Wilce, M.C.(2000) Nat Struct Biol 7: 1051-1054
- PubMed: 11062562
- DOI: https://doi.org/10.1038/80975
- Primary Citation of Related Structures:
1FS0 - PubMed Abstract:
ATP synthases (F(1)F(o)-ATPases) use energy released by the movement of protons down a transmembrane electrochemical gradient to drive the synthesis of ATP, the universal biological energy currency. Proton flow through F(o) drives rotation of a ring of c-subunits and a complex of the gamma and epsilon-subunits, causing cyclical conformational changes in F(1) that are required for catalysis. The crystal structure of a large portion of F(1) has been resolved. However, the structure of the central portion of the enzyme, through which conformational changes in F(o) are communicated to F(1), has until now remained elusive. Here we report the crystal structure of a complex of the epsilon-subunit and the central domain of the gamma-subunit refined at 2.1 A resolution. The structure reveals how rotation of these subunits causes large conformational changes in F(1), and thereby provides new insights into energy coupling between F(o) and F(1).
Organizational Affiliation:
Crystallography Centre Department of Pharmacology, University of Western Australia and Western Australian Institute for Medical Research, Nedlands Western Australia 6907, Australia.