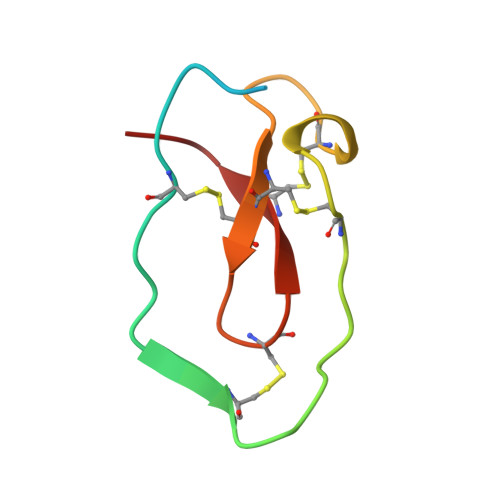
Crystal structure of an elastase-specific inhibitor elafin complexed with porcine pancreatic elastase determined at 1.9 A resolution.
Tsunemi, M., Matsuura, Y., Sakakibara, S., Katsube, Y.(1996) Biochemistry 35: 11570-11576
- PubMed: 8794736
- DOI: https://doi.org/10.1021/bi960900l
- Primary Citation of Related Structures:
1FLE - PubMed Abstract:
The crystal structure of a stoichiometric complex between an elastase-specific inhibitor elafin and porcine pancreatic elastase (PPE) has been determined and refined to a crystallographic R-factor of 19.7% at 1.9 A resolution. The polypeptide chain of elafin has a planar spiral shape with an exposed external part and an internal core part which resembles both the crystal structure of human seminal plasma inhibitor (HUSI-1) [Grütter, M. G., Fendrich, G., Huber, R., & Bode, W. (1988) EMBO J. 7, 345-351] and the solution structure of Na+,K(+)-ATPase inhibitor (SPAI-1) revealed by NMR analysis [Kozaki, T., Kawakami, Y., Tachibana, S., Hatanaka, H., & Inagaki, F. (1994) Pept. Chem., 405-408]. The external region containing the primary binding loop is interconnected by four disulfide bonds to the internal part composed of a beta-sheet and a hairpin loop. The scissile peptide bond Ala24i(P1)-Met25i(P1') in the primary binding site is intact, and its carbonyl carbon is in van der Waals contact with O gamma of the active site Ser195 of PPE. The seven residues of Leu20i(P5)-Leu26i(P2') of the primary binding loop and the three residues of Ser48i, Cys49i, and Ala52i of the adjacent hairpin loop are in contact with PPE by hydrogen bonds and/or van der Waals interactions in a manner similar to that observed for other serine protease-inhibitor complexes. Electron densities of the N-terminal residues Ala1i-Ser10i which are not responsible for the elastase inhibitory activity were not visible, probably due to disordered conformation. The guanido group (N eta 1, N eta 2) of Arg61 in the complex interacts with S delta of Met25i(P1') by possible hydrogen bonds between N and S atoms, accompanying a large positional shift of the side chain of Arg61-(S1') between the complexed and free forms of PPE. The primary binding site is stabilized by hydrogen bonds between the guanido group (N eta 1, N eta 2) of Arg22i(P3) and the carbonyl group of Met25i(P1') across the scissile bond, as well as by a hydrogen bond between the amino group of Cys23i(P2) and the carbonyl group of Ser48i in the internal core. This intramolecular hydrogen bond network and the network of four disulfide bonds might play a significant role in stabilizing the conformation of the binding site for expressing the potent specific inhibitory activity.
Organizational Affiliation:
Peptide Institute Inc., Protein Research Foundation, Osaka, Japan.