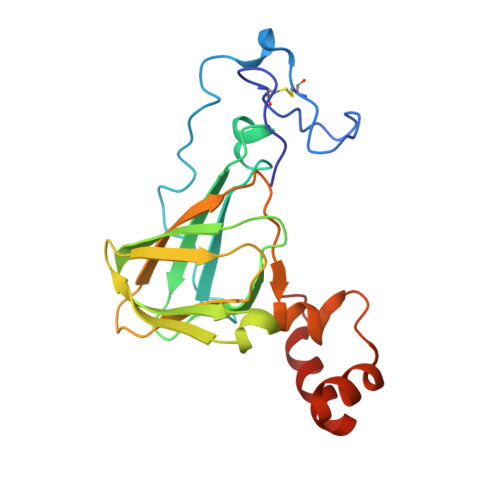
Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities.
Woo, E.J., Dunwell, J.M., Goodenough, P.W., Marvier, A.C., Pickersgill, R.W.(2000) Nat Struct Biol 7: 1036-1040
- PubMed: 11062559
- DOI: https://doi.org/10.1038/80954
- Primary Citation of Related Structures:
1FI2 - PubMed Abstract:
Germin is a hydrogen peroxide generating oxalate oxidase with extreme thermal stability; it is involved in the defense against biotic and abiotic stress in plants. The structure, determined at 1.6 A resolution, comprises beta-jellyroll monomers locked into a homohexamer (a trimer of dimers), with extensive surface burial accounting for its remarkable stability. The germin dimer is structurally equivalent to the monomer of the 7S seed storage proteins (vicilins), indicating evolution from a common ancestral protein. A single manganese ion is bound per germin monomer by ligands similar to those of manganese superoxide dismutase (MnSOD). Germin is also shown to have SOD activity and we propose that the defense against extracellular superoxide radicals is an important additional role for germin and related proteins.
Organizational Affiliation:
Institute of Food Research, Reading Laboratory, Earley Gate, Reading RG6 6BZ, UK.