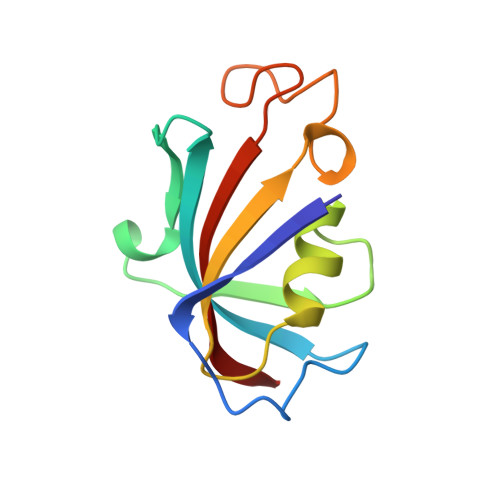
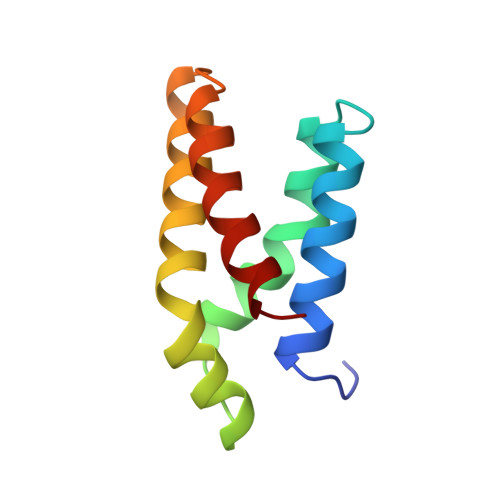
Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP.
Choi, J., Chen, J., Schreiber, S.L., Clardy, J.(1996) Science 273: 239-242
- PubMed: 8662507
- DOI: https://doi.org/10.1126/science.273.5272.239
- Primary Citation of Related Structures:
1FAP - PubMed Abstract:
Rapamycin, a potent immunosuppressive agent, binds two proteins: the FK506-binding protein (FKBP12) and the FKBP-rapamycin-associated protein (FRAP). A crystal structure of the ternary complex of human FKBP12, rapamycin, and the FKBP12-rapamycin-binding (FRB) domain of human FRAP at a resolution of 2.7 angstroms revealed the two proteins bound together as a result of the ability of rapamycin to occupy two different hydrophobic binding pockets simultaneously. The structure shows extensive interactions between rapamycin and both proteins, but fewer interactions between the proteins. The structure of the FRB domain of FRAP clarifies both rapamycin-independent and -dependent effects observed for mutants of FRAP and its homologs in the family of proteins related to the ataxia-telangiectasia mutant gene product, and it illustrates how a small cell-permeable molecule can mediate protein dimerization.
Organizational Affiliation:
Department of Chemistry, Baker Laboratory, Cornell University, Ithaca, NY 14853-1301, USA.