NMR structure of the N-terminal J domain of murine polyomavirus T antigens. Implications for DnaJ-like domains and for mutations of T antigens.
Berjanskii, M.V., Riley, M.I., Xie, A., Semenchenko, V., Folk, W.R., Van Doren, S.R.(2000) J Biol Chem 275: 36094-36103
- PubMed: 10950962
- DOI: https://doi.org/10.1074/jbc.M006572200
- Primary Citation of Related Structures:
1FAF - PubMed Abstract:
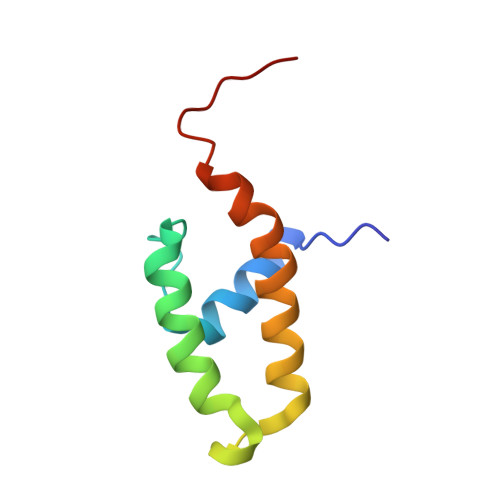
The NMR structure of the N-terminal, DnaJ-like domain of murine polyomavirus tumor antigens (PyJ) has been determined to high precision, with root mean square deviations to the mean structure of 0.38 A for backbone atoms and 0.94 A for all heavy atoms of ordered residues 5-41 and 50-69. PyJ possesses a three-helix fold, in which anti-parallel helices II and III are bridged by helix I, similar to the four-helix fold of the J domains of DnaJ and human DnaJ-1. PyJ differs significantly in the lengths of N terminus, helix I, and helix III. The universally conserved HPD motif appears to form a His-Pro C-cap of helix II. Helix I features a stabilizing Schellman C-cap that is probably conserved universally among J domains. On the helix II surface where positive charges of other J domains have been implicated in binding of hsp70s, PyJ contains glutamine residues. Nonetheless, chimeras that replace the J domain of DnaJ with PyJ function like wild-type DnaJ in promoting growth of Escherichia coli. This activity can be modulated by mutations of at least one of these glutamines. T antigen mutations reported to impair cellular transformation by the virus, presumably via interactions with PP2A, cluster in the hydrophobic folding core and at the extreme N terminus, remote from the HPD loop.
Organizational Affiliation:
Department of Biochemistry, University of Missouri, Columbia, Missouri 65211, USA.