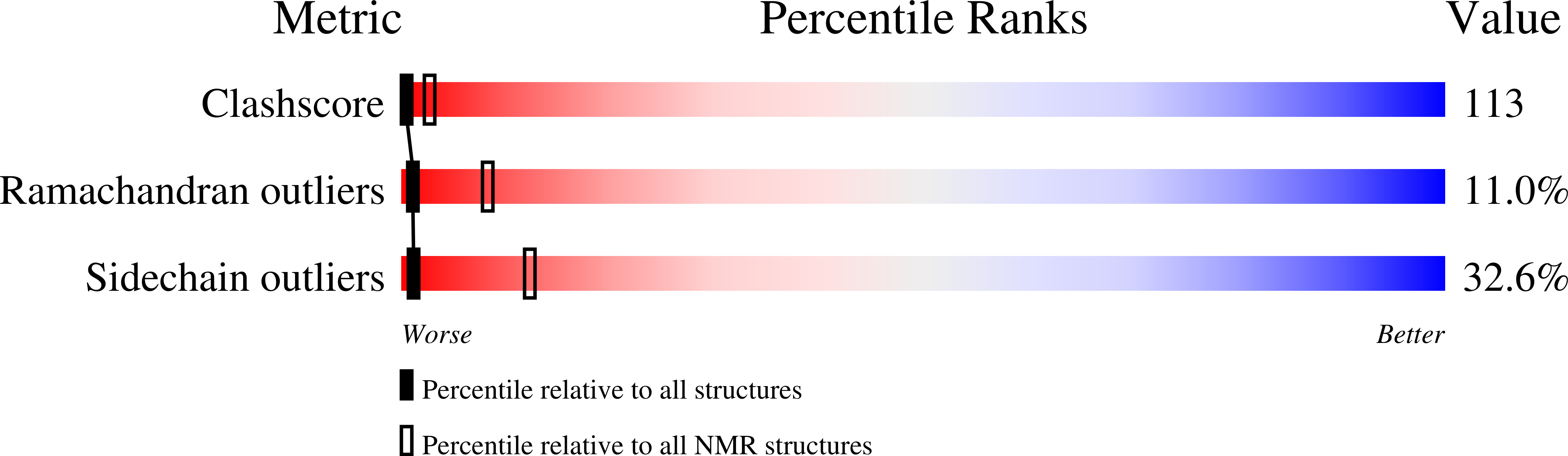Three-dimensional solution structure of omega-conotoxin TxVII, an L-type calcium channel blocker.
Kobayashi, K., Sasaki, T., Sato, K., Kohno, T.(2000) Biochemistry 39: 14761-14767
- PubMed: 11101291
- DOI: https://doi.org/10.1021/bi001506x
- Primary Citation of Related Structures:
1F3K - PubMed Abstract:
We determined the three-dimensional structure of omega-conotoxin TxVII, a 26-residue peptide that is an L-type calcium channel blocker, by (1)H NMR in aqueous solution. Twenty converged structures of this peptide were obtained on the basis of 411 distance constraints obtained from nuclear Overhauser effect connectivities, 20 torsion angle constraints, and 21 constraints associated with hydrogen bonds and disulfide bonds. The root-mean-square deviations about the averaged coordinates of the backbone atoms (N, C(alpha), C, and O) and all heavy atoms were 0.50 +/- 0.09 A and 0.99 +/- 0.13 A, respectively. The structure of omega-conotoxin TxVII is composed of a triple-stranded antiparallel beta-sheet and four turns. The three disulfide bonds in omega-conotoxin TxVII form the classical cystine knot motif of toxic or inhibitory polypeptides. The overall folding of omega-conotoxin TxVII is similar to those of the N-type calcium channel blockers, omega-conotoxin GVIA and MVIIA, despite the low amino acid sequence homology among them. omega-Conotoxin TxVII exposes many hydrophobic residues to a certain surface area. In contrast, omega-conotoxin GVIA and MVIIA expose basic residues in the same way as omega-conotoxin TxVII. The channel binding site of omega-conotoxin TxVII is different from those of omega-conotoxin GVIA and MVIIA, although the overall folding of these three peptides is similar. The gathered hydrophobic residues of omega-conotoxin TxVII probably interact with the hydrophobic cluster of the alpha(1) subunit of the L-type calcium channel, which consists of 13 residues located in segments 5 and 6 in domain III and in segment 6 in domain IV.
Organizational Affiliation:
Mitsubishi Kasei Institute of Life Sciences, Minamiooya, Machida, Tokyo 194-8511, Japan.