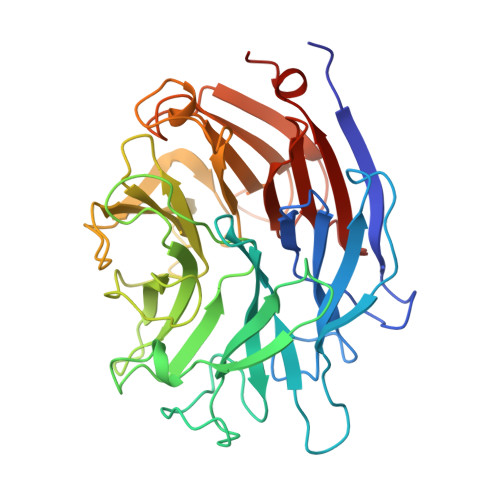
The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Gaskell, A., Crennell, S., Taylor, G.(1995) Structure 3: 1197-1205
- PubMed: 8591030
- DOI: https://doi.org/10.1016/s0969-2126(01)00255-6
- Primary Citation of Related Structures:
1EUR, 1EUS, 1EUT - PubMed Abstract:
Sialidases, or neuraminidases, have been implicated in the pathogenesis of many diseases, but are also produced by many non-pathogenic bacteria. Bacterial sialidases are very variable in size, often possessing domains in addition to the catalytic domain. The sialidase from the non-pathogenic soil bacterium Micromonospora viridifaciens is secreted in two forms with molecular weights of 41 kDa or 68 kDa, depending on the nature of the carbohydrate used to induce expression.
Organizational Affiliation:
School of Biology and Biochemistry, University of Bath, UK.