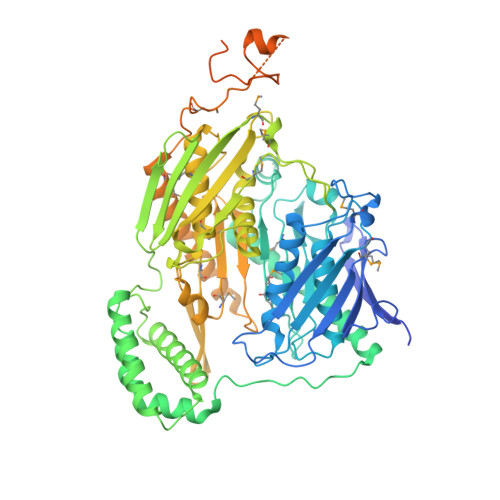
A Duplicated Fold is the Structural Basis for Polynucleotide Phosphorylase Catalytic Activity, Processivity, and Regulation
Symmons, M.F., Jones, G.H., Luisi, B.F.(2000) Structure 8: 1215
- PubMed: 11080643
- DOI: https://doi.org/10.1016/s0969-2126(00)00521-9
- Primary Citation of Related Structures:
1E3H, 1E3P - PubMed Abstract:
Polynucleotide phosphorylase (PNPase) is a polyribonucleotide nucleotidyl transferase (E.C.2.7.7.8) that degrades mRNA in prokaryotes. Streptomyces antibioticus PNPase also assays as a guanosine 3'-diphosphate 5'-triphosphate (pppGpp) synthetase (E.C.2.7.6.5). It may function to coordinate changes in mRNA lifetimes with pppGpp levels during the Streptomyces lifecycle.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom. mfs@mole.bio.cam.ac.uk