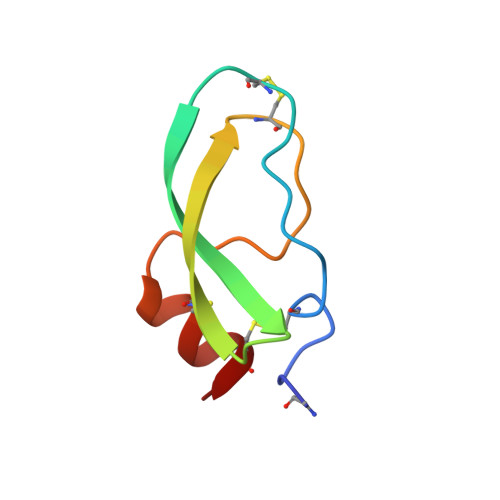
Crystal structure of alpha-dendrotoxin from the green mamba venom and its comparison with the structure of bovine pancreatic trypsin inhibitor.
Skarzynski, T.(1992) J Mol Biol 224: 671-683
- PubMed: 1373774
- DOI: https://doi.org/10.1016/0022-2836(92)90552-u
- Primary Citation of Related Structures:
1DTX - PubMed Abstract:
The three-dimensional structure of alpha-dendrotoxin (alpha-DTX) from the green mamba (Dendroaspis angusticeps) venom has been determined crystallographically using the method of isomorphous replacement and refined at 2.2 A resolution using a restrained least-squares method. The crystallographic R-factor is 0.169 for all 3451 measured reflections between 7.0 and 2.2 A. Although the main-chain fold of alpha-DTX is similar to that of homologous bovine pancreatic trypsin inhibitor (BPTI), there are significant differences involving segments of the polypeptide chain close to the "antiprotease site" of BPTI. Comparison of the structure of alpha-DTX with the existing models of BPTI and its complexes with trypsin and kallikrein reveals structural differences that explain the inability of alpha-DTX to inhibit trypsin and chymotrypsin.
Organizational Affiliation:
Blackett Laboratory, Imperial College, London, England.