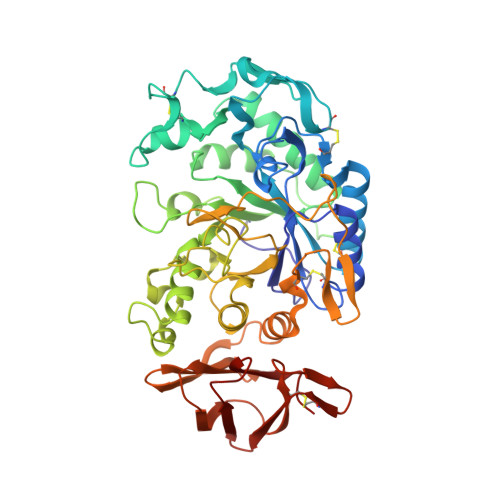
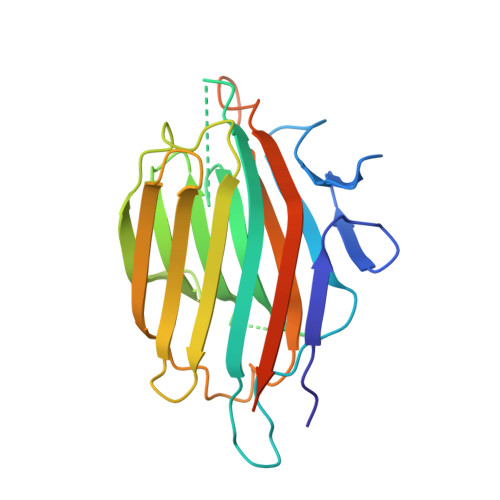
Substrate mimicry in the active center of a mammalian alpha-amylase: structural analysis of an enzyme-inhibitor complex.
Bompard-Gilles, C., Rousseau, P., Rouge, P., Payan, F.(1996) Structure 4: 1441-1452
- PubMed: 8994970
- DOI: https://doi.org/10.1016/s0969-2126(96)00151-7
- Primary Citation of Related Structures:
1DHK - PubMed Abstract:
alpha-Amylases catalyze the hydrolysis of glycosidic linkages in starch and other related polysaccharides. The alpha-amylase inhibitor (alpha-Al) from the bean Phaseolus vulgaris belongs to a family of plant defence proteins and is a potent inhibitor of mammalian alpha-amylases. The structure of pig pancreatic alpha-amylase (PPA) in complex with both a carbohydrate inhibitor (acarbose) and a proteinaceous inhibitor (Tendamistat) is known, but the catalytic mechanism is poorly understood.
Organizational Affiliation:
AFMB-IBSM-CNRS, Marseille, France.