Structure of carboxypeptidase B at 2-8 A resolution.
Schmid, M.F., Herriott, J.R.(1976) J Mol Biol 103: 175-190
- PubMed: 957425
- DOI: https://doi.org/10.1016/0022-2836(76)90058-9
- Primary Citation of Related Structures:
1CPB
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(1976) J Mol Biol 103: 175-190
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CARBOXYPEPTIDASE B | 82 | Bos taurus | Mutation(s): 0 EC: 3.4.17.2 | 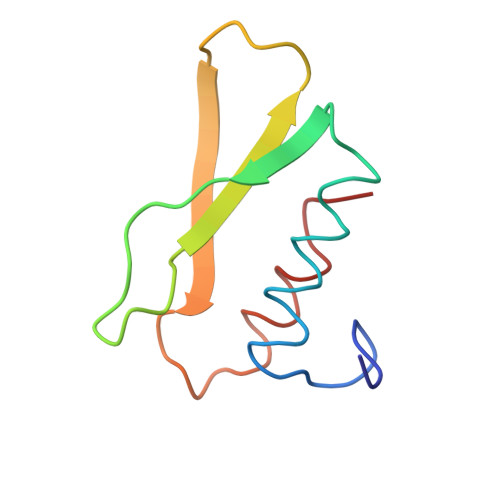 | |
UniProt | |||||
Find proteins for P00732 (Bos taurus) Explore P00732 Go to UniProtKB: P00732 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00732 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CARBOXYPEPTIDASE B | 217 | Bos taurus | Mutation(s): 0 EC: 3.4.17.2 | 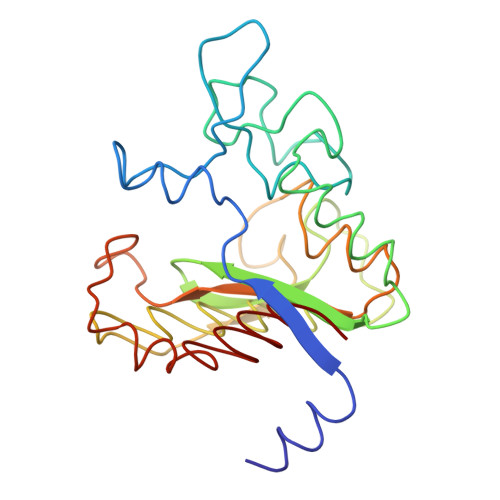 | |
UniProt | |||||
Find proteins for P00732 (Bos taurus) Explore P00732 Go to UniProtKB: P00732 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00732 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 54.94 | α = 90 |
| b = 54.94 | β = 90 |
| c = 104.6 | γ = 120 |