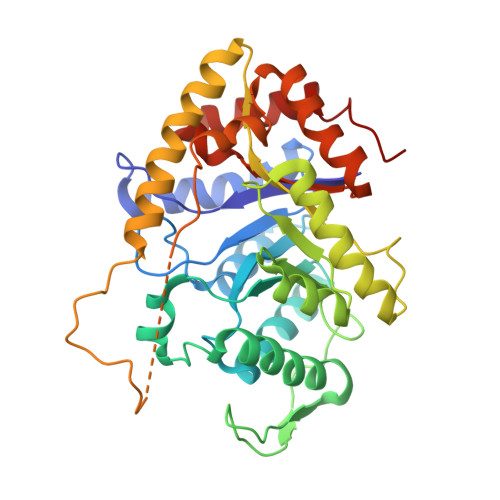
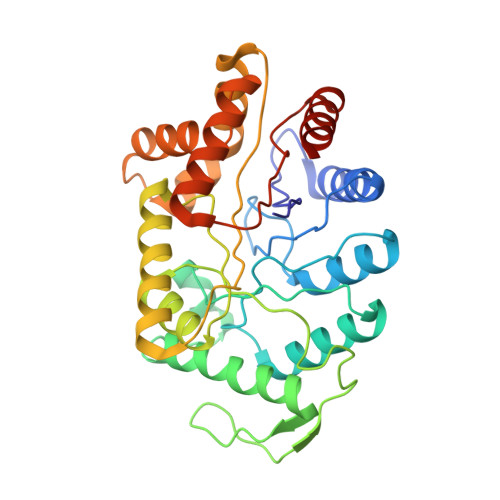
Three-dimensional structure of bacterial luciferase from Vibrio harveyi at 2.4 A resolution.
Fisher, A.J., Raushel, F.M., Baldwin, T.O., Rayment, I.(1995) Biochemistry 34: 6581-6586
- PubMed: 7756289
- DOI: https://doi.org/10.1021/bi00020a002
- Primary Citation of Related Structures:
1BRL - PubMed Abstract:
Luciferases are a class of enzymes that generate light in the visible spectrum. Luciferase from luminous marine bacteria is an alpha-beta heterodimer monooxygenase that catalyzes the oxidation of FMNH2 and a long-chain aliphatic aldehyde. The X-ray crystal structure of bacterial luciferase from Vibrio harveyi has been determined to 2.4 A resolution. The structure was solved by a combination of multiple isomorphous replacement and molecular averaging between the two heterodimers in the asymmetric unit. Each subunit folds into a (beta/alpha)8 barrel motif, and dimerization is mediated through a parallel four-helix bundle centered on a pseudo 2-fold axis that relates the structurally similar subunits. The vicinity of the active site has been identified on the alpha subunit by correlations with similar protein motifs and previous biochemical studies. The structure presented here represents the first molecular model of a bioluminescent enzyme.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin, Madison 53705, USA.