Structure of the IGF-binding domain of the insulin-like growth factor-binding protein-5 (IGFBP-5): implications for IGF and IGF-I receptor interactions.
Kalus, W., Zweckstetter, M., Renner, C., Sanchez, Y., Georgescu, J., Grol, M., Demuth, D., Schumacher, R., Dony, C., Lang, K., Holak, T.A.(1998) EMBO J 17: 6558-6572
- PubMed: 9822601
- DOI: https://doi.org/10.1093/emboj/17.22.6558
- Primary Citation of Related Structures:
1BOE - PubMed Abstract:
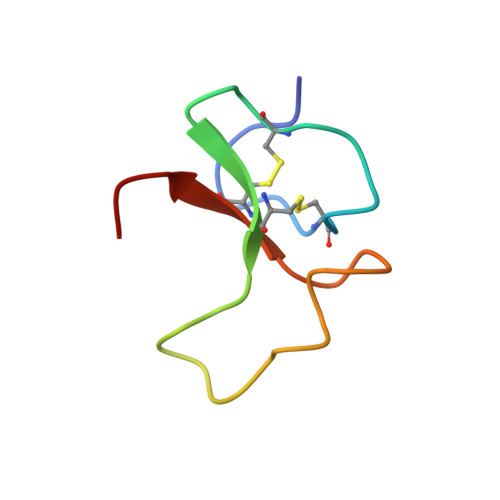
Binding proteins for insulin-like growth factors (IGFs) IGF-I and IGF-II, known as IGFBPs, control the distribution, function and activity of IGFs in various cell tissues and body fluids. Insulin-like growth factor-binding protein-5 (IGFBP-5) is known to modulate the stimulatory effects of IGFs and is the major IGF-binding protein in bone tissue. We have expressed two N-terminal fragments of IGFBP-5 in Escherichia coli; the first encodes the N-terminal domain of the protein (residues 1-104) and the second, mini-IGFBP-5, comprises residues Ala40 to Ile92. We show that the entire IGFBP-5 protein contains only one high-affinity binding site for IGFs, located in mini-IGFBP-5. The solution structure of mini-IGFBP-5, determined by nuclear magnetic resonance spectroscopy, discloses a rigid, globular structure that consists of a centrally located three-stranded anti-parallel beta-sheet. Its scaffold is stabilized further by two inside packed disulfide bridges. The binding to IGFs, which is in the nanomolar range, involves conserved Leu and Val residues localized in a hydrophobic patch on the surface of the IGFBP-5 protein. Remarkably, the IGF-I receptor binding assays of IGFBP-5 showed that IGFBP-5 inhibits the binding of IGFs to the IGF-I receptor, resulting in reduction of receptor stimulation and autophosphorylation. Compared with the full-length IGFBP-5, the smaller N-terminal fragments were less efficient inhibitors of the IGF-I receptor binding of IGFs.
Organizational Affiliation:
Max Planck Institute for Biochemistry, D-82152 Martinsried.