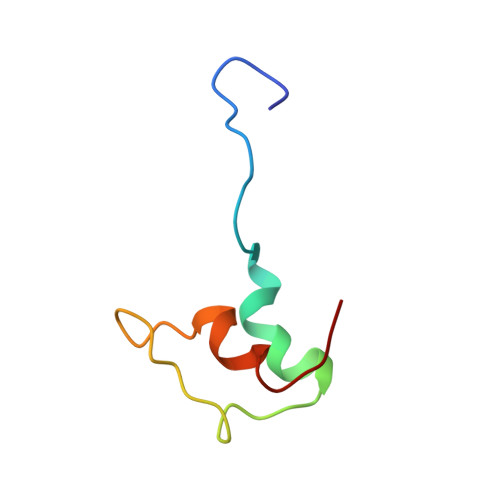
Three-dimensional solution structure of the E3-binding domain of the dihydrolipoamide succinyltransferase core from the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli.
Robien, M.A., Clore, G.M., Omichinski, J.G., Perham, R.N., Appella, E., Sakaguchi, K., Gronenborn, A.M.(1992) Biochemistry 31: 3463-3471
- PubMed: 1554728
- DOI: https://doi.org/10.1021/bi00128a021
- Primary Citation of Related Structures:
1BAL, 1BBL - PubMed Abstract:
The three-dimensional solution structure of a 51-residue synthetic peptide comprising the dihydrolipoamide dehydrogenase (E3)-binding domain of the dihydrolipoamide succinyltransferase (E2) core of the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli has been determined by nuclear magnetic resonance spectroscopy and hybrid distance geometry-dynamical simulated annealing calculations. The structure is based on 630 approximate interproton distance and 101 torsion angle (phi, psi, chi 1) restraints. A total of 56 simulated annealing structures were calculated, and the atomic rms distribution about the mean coordinate positions for residues 12-48 of the synthetic peptide is 1.24 A for the backbone atoms, 1.68 A for all atoms, and 1.33 A for all atoms excluding the six side chains which are disordered at chi 1 and the seven which are disordered at chi 2; when the irregular partially disordered loop from residues 31 to 39 is excluded, the rms distribution drops to 0.77 A for the backbone atoms, 1.55 A for all atoms, and 0.89 A for ordered side chains. Although proton resonance assignments for the N-terminal 11 residues and the C-terminal 3 residues were obtained, these two segments of the polypeptide are disordered in solution as evidenced by the absence of nonsequential nuclear Overhauser effects. The solution structure of the E3-binding domain consists of two parallel helices (residues 14-23 and 40-48), a short extended strand (24-26), a five-residue helical-like turn, and an irregular (and more disordered) loop (residues 31-39). This report presents the first structure of an E3-binding domain from a 2-oxo acid dehydrogenase complex.(ABSTRACT TRUNCATED AT 250 WORDS)
Organizational Affiliation:
Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892.