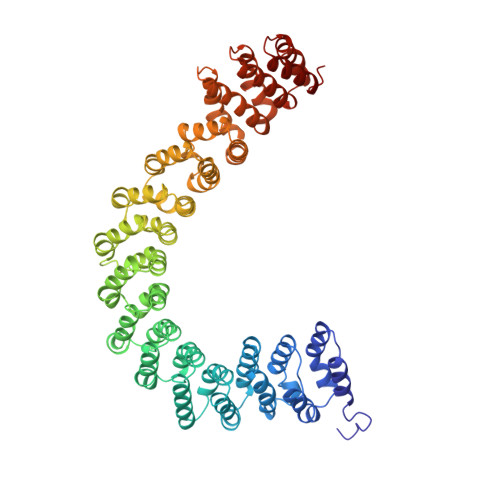The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs.
Groves, M.R., Hanlon, N., Turowski, P., Hemmings, B.A., Barford, D.(1999) Cell 96: 99-110
- PubMed: 9989501
- DOI: https://doi.org/10.1016/s0092-8674(00)80963-0
- Primary Citation of Related Structures:
1B3U - PubMed Abstract:
The PR65/A subunit of protein phosphatase 2A serves as a scaffolding molecule to coordinate the assembly of the catalytic subunit and a variable regulatory B subunit, generating functionally diverse heterotrimers. Mutations of the beta isoform of PR65 are associated with lung and colon tumors. The crystal structure of the PR65/Aalpha subunit, at 2.3 A resolution, reveals the conformation of its 15 tandemly repeated HEAT sequences, degenerate motifs of approximately 39 amino acids present in a variety of proteins, including huntingtin and importin beta. Individual motifs are composed of a pair of antiparallel alpha helices that assemble in a mainly linear, repetitive fashion to form an elongated molecule characterized by a double layer of alpha helices. Left-handed rotations at three interrepeat interfaces generate a novel left-hand superhelical conformation. The protein interaction interface is formed from the intrarepeat turns that are aligned to form a continuous ridge.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, United Kingdom.