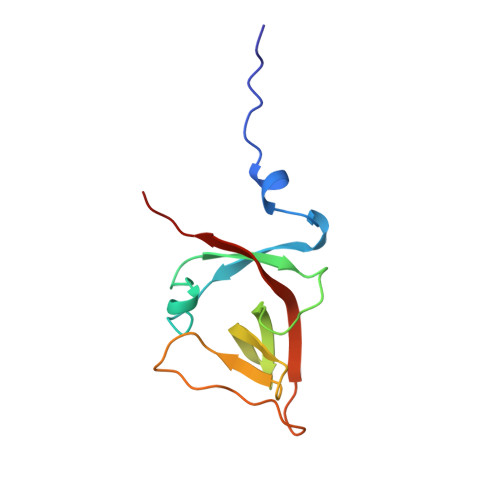
The UmuD' protein filament and its potential role in damage induced mutagenesis.
Peat, T.S., Frank, E.G., McDonald, J.P., Levine, A.S., Woodgate, R., Hendrickson, W.A.(1996) Structure 4: 1401-1412
- PubMed: 8994967
- DOI: https://doi.org/10.1016/s0969-2126(96)00148-7
- Primary Citation of Related Structures:
1AY9 - PubMed Abstract:
Damage induced 'SOS mutagenesis' may occur transiently as part of the global SOS response to DNA damage in bacteria. A key participant in this process is the UmuD protein, which is produced in an inactive from but converted to the active form, UmuD', by a RecA-mediated self-cleavage reaction. UmuD', together with UmuC and activated RecA (RecA*), enables the DNA polymerase III holoenzyme to replicate across chemical and UV induced lesions. The efficiency of this reaction depends on several intricate protein-protein interactions.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA.