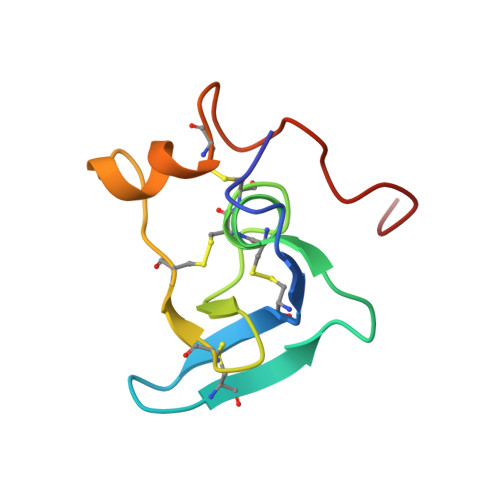
Solution structure of the transforming growth factor beta-binding protein-like module, a domain associated with matrix fibrils.
Yuan, X., Downing, A.K., Knott, V., Handford, P.A.(1997) EMBO J 16: 6659-6666
- PubMed: 9362480
- DOI: https://doi.org/10.1093/emboj/16.22.6659
- Primary Citation of Related Structures:
1APJ - PubMed Abstract:
Here we describe the high resolution nuclear magnetic resonance (NMR) structure of a transforming growth factor beta (TGF-beta)-binding protein-like (TB) domain, which comes from human fibrillin-1, the protein defective in the Marfan syndrome (MFS). This domain is found in fibrillins and latent TGF-beta-binding proteins (LTBPs) which are localized to fibrillar structures in the extracellular matrix. The TB domain manifests a novel fold which is globular and comprises six antiparallel beta-strands and two alpha-helices. An unusual cysteine triplet conserved in the sequences of TB domains is localized to the hydrophobic core, at the C-terminus of an alpha-helix. The structure is stabilized by four disulfide bonds which pair in a 1-3, 2-6, 4-7, 5-8 pattern, two of which are solvent exposed. Analyses of MFS-causing mutations and the fibrillin-1 cell-binding RGD site provide the first clues to the surface specificity of TB domain interactions. Modelling of a homologous TB domain from LTBP-1 (residues 1018-1080) suggests that hydrophobic contacts may play a role in its interaction with the TGF-beta1 latency-associated peptide.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, Oxford OX1 3QU, UK.