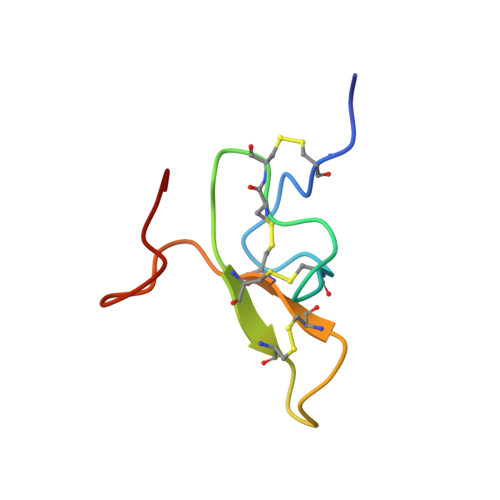
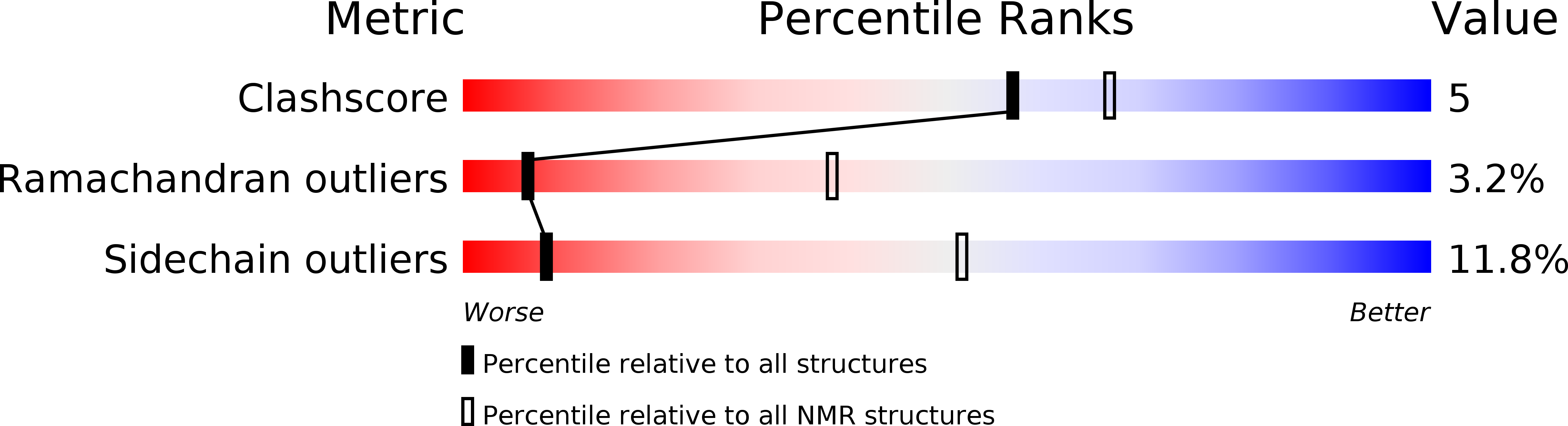The solution structure of omega-Aga-IVB, a P-type calcium channel antagonist from venom of the funnel web spider, Agelenopsis aperta.
Reily, M.D., Thanabal, V., Adams, M.E.(1995) J Biomol NMR 5: 122-132
- PubMed: 7703698
- DOI: https://doi.org/10.1007/BF00208803
- Primary Citation of Related Structures:
1AGG - PubMed Abstract:
The 48 amino acid peptides omega-Aga-IVA and omega-Aga-IVB are the first agents known to specifically block P-type calcium channels in mammalian brain, thus complementing the existing suite of pharmacological tools used for characterizing calcium channels. These peptides provide a new set of probes for studies aimed at elucidating the structural basis underlying the subtype specificity of calcium channel antagonists. We used 288 NMR-derived constraints in a protocol combining distance geometry and molecular dynamics employing the program DGII, followed by energy minimization with Discover to derive the three-dimensional structure of omega-Aga-IVB. The toxin consists of a well-defined core region, comprising seven solvent-shielded residues and a well-defined triple-stranded beta-sheet. Four loop regions have average backbone rms deviations between 0.38 and 1.31 A, two of which are well-defined type-II beta-turns. Other structural features include disordered C- and N-termini and several conserved basic amino acids that are clustered on one face of the molecule. The reported structure suggests a possible surface for interaction with the channel. This surface contains amino acids that are identical to those of another known P-type calcium channel antagonist, omega-Aga-IVA, and is rich in basic residues that may have a role in binding to the anionic sites in the extracellular regions of the calcium channel.
Organizational Affiliation:
Department of Chemistry, Parke-Davis Pharmaceutical Research, Division of Warner Lambert Company, Ann Arbor, MI 48105.