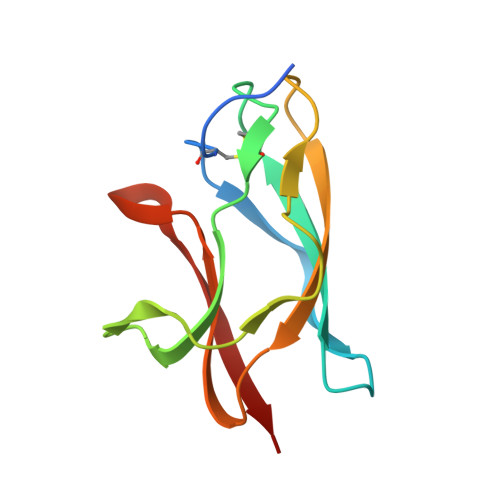
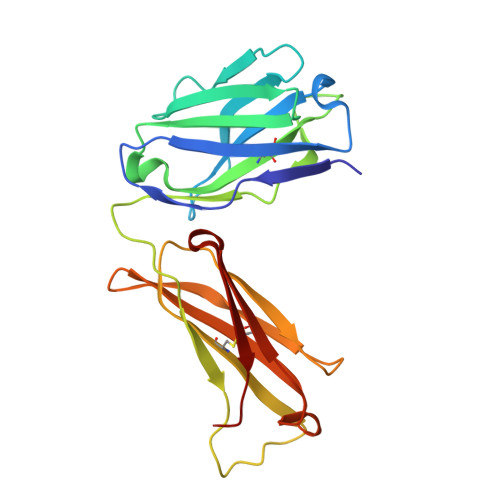
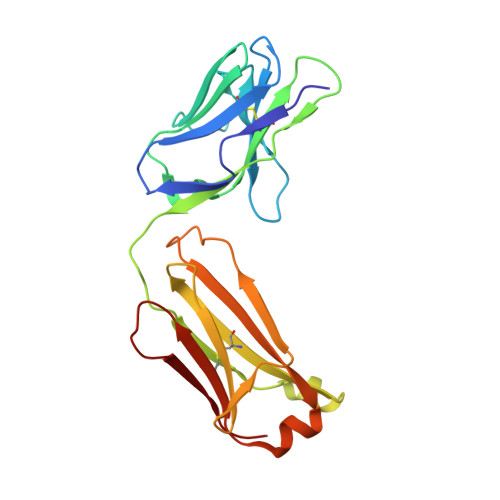
Structural basis of West Nile virus neutralization by a therapeutic antibody.
Nybakken, G.E., Oliphant, T., Johnson, S., Burke, S., Diamond, M.S., Fremont, D.H.(2005) Nature 437: 764-769
- PubMed: 16193056
- DOI: https://doi.org/10.1038/nature03956
- Primary Citation of Related Structures:
1ZTX - PubMed Abstract:
West Nile virus is a mosquito-borne flavivirus closely related to the human epidemic-causing dengue, yellow fever and Japanese encephalitis viruses. In establishing infection these icosahedral viruses undergo endosomal membrane fusion catalysed by envelope glycoprotein rearrangement of the putative receptor-binding domain III (DIII) and exposure of the hydrophobic fusion loop. Humoral immunity has an essential protective function early in the course of West Nile virus infection. Here, we investigate the mechanism of neutralization by the E16 monoclonal antibody that specifically binds DIII. Structurally, the E16 antibody Fab fragment engages 16 residues positioned on four loops of DIII, a consensus neutralizing epitope sequence conserved in West Nile virus and distinct in other flaviviruses. The E16 epitope protrudes from the surface of mature virions in three distinct environments, and docking studies predict Fab binding will leave five-fold clustered epitopes exposed. We also show that E16 inhibits infection primarily at a step after viral attachment, potentially by blocking envelope glycoprotein conformational changes. Collectively, our results suggest that a vaccine strategy targeting the dominant DIII epitope may elicit safe and effective immune responses against flaviviral diseases.
Organizational Affiliation:
Department of Pathology, Washington University School of Medicine, St Louis, Missouri 63110, USA.