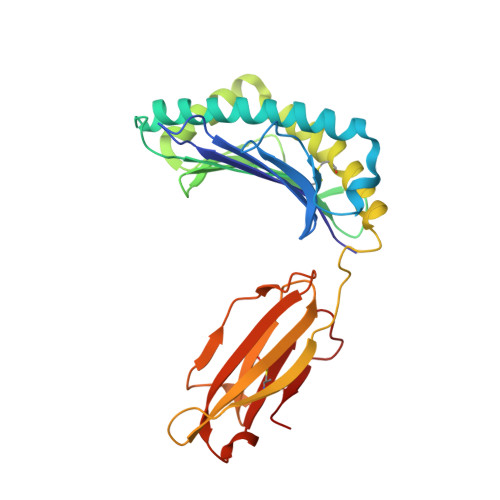
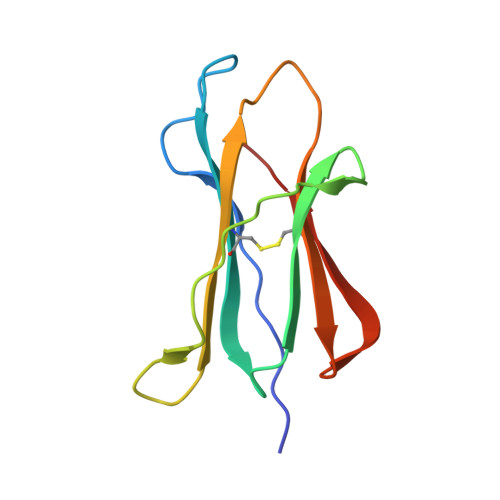
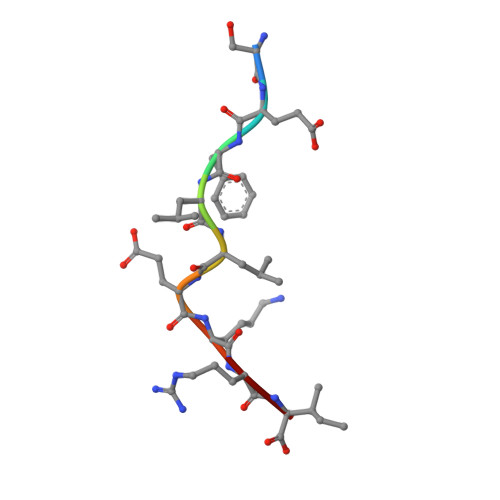
The H-2Kk MHC peptide-binding groove anchors the backbone of an octameric antigenic peptide in an unprecedented mode.
Kellenberger, C., Roussel, A., Malissen, B.(2005) J Immunol 175: 3819-3825
- PubMed: 16148128
- DOI: https://doi.org/10.4049/jimmunol.175.6.3819
- Primary Citation of Related Structures:
1ZT1, 1ZT7 - PubMed Abstract:
A wealth of data has accumulated on the structure of mouse MHC class I (MHCI) molecules encoded by the H-2(b) and H-2(d) haplotypes. In contrast, there is a dearth of structural data regarding H-2(k)-encoded molecules. Therefore, the structures of H-2K(k) complexed to an octameric peptide from influenza A virus (HA(259-266)) and to a nonameric peptide from SV40 (SV40(560-568)) have been determined by x-ray crystallography at 2.5 and 3.0 A resolutions, respectively. The structure of the H-2K(k)-HA(259-266) complex reveals that residues located on the floor of the peptide-binding groove contact directly the backbone of the octameric peptide and force it to lie deep within the H-2K(k) groove. This unprecedented mode of peptide binding occurs despite the presence of bulky residues in the middle of the floor of the H-2K(k) peptide-binding groove. As a result, the Calpha atoms of peptide residues P5 and P6 are more buried than the corresponding residues of H-2K(b)-bound octapeptides, making them even less accessible to TCR contact. When bound to H-2K(k), the backbone of the SV40(560-568) nonapeptide bulges out of the peptide-binding groove and adopts a conformation reminiscent of that observed for peptides bound to H-2L(d). This structural convergence occurs despite the totally different architectures of the H-2L(d) and H-2K(k) peptide-binding grooves. Therefore, these two H-2K(k)-peptide complexes provide insights into the mechanisms through which MHC polymorphism outside primary peptide pockets influences the conformation of the bound peptides and have implications for TCR recognition and vaccine design.
Organizational Affiliation:
Centre d'Immunologie de Marseille-Luminy, Institut National de la Santé et de la Recherche Médicale-Centre National Recherche de la Scientifique-Université de la Méditerranée, Parc Scientifique de Luminy, Marseille, France. kellenberger@ciml.univ-mrs.fr