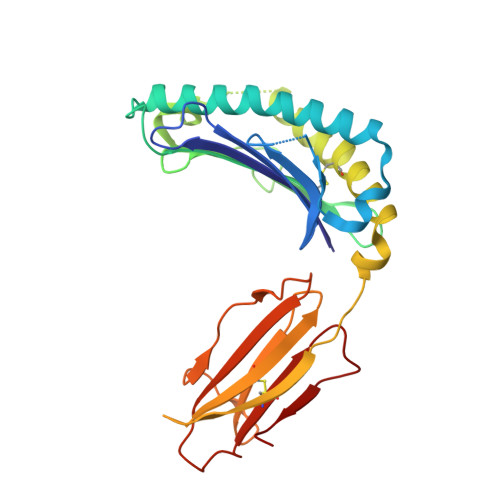
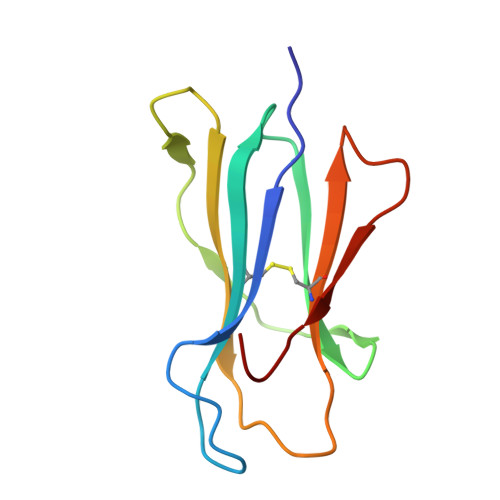
Structure of a pheromone receptor-associated MHC molecule with an open and empty groove.
Olson, R., Huey-Tubman, K.E., Dulac, C., Bjorkman, P.J.(2005) PLoS Biol 3: e257-e257
- PubMed: 16089503
- DOI: https://doi.org/10.1371/journal.pbio.0030257
- Primary Citation of Related Structures:
1ZS8 - PubMed Abstract:
Neurons in the murine vomeronasal organ (VNO) express a family of class Ib major histocompatibility complex (MHC) proteins (M10s) that interact with the V2R class of VNO receptors. This interaction may play a direct role in the detection of pheromonal cues that initiate reproductive and territorial behaviors. The crystal structure of M10.5, an M10 family member, is similar to that of classical MHC molecules. However, the M10.5 counterpart of the MHC peptide-binding groove is open and unoccupied, revealing the first structure of an empty class I MHC molecule. Similar to empty MHC molecules, but unlike peptide-filled MHC proteins and non-peptide-binding MHC homologs, M10.5 is thermally unstable, suggesting that its groove is normally occupied. However, M10.5 does not bind endogenous peptides when expressed in mammalian cells or when offered a mixture of class I-binding peptides. The F pocket side of the M10.5 groove is open, suggesting that ligands larger than 8-10-mer class I-binding peptides could fit by extending out of the groove. Moreover, variable residues point up from the groove helices, rather than toward the groove as in classical MHC structures. These data suggest that M10s are unlikely to provide specific recognition of class I MHC-binding peptides, but are consistent with binding to other ligands, including proteins such as the V2Rs.
Organizational Affiliation:
Division of Biology, California Institute of Technology, Pasadena, California, USA.