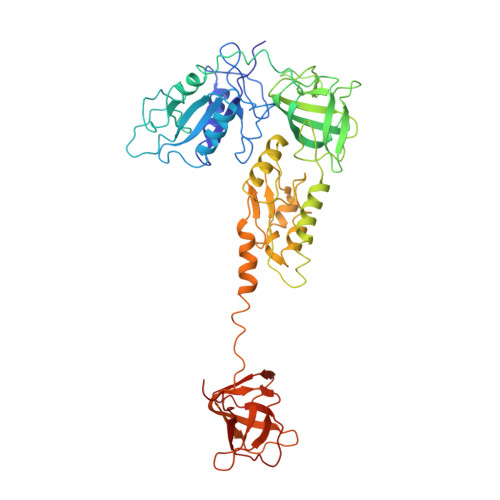
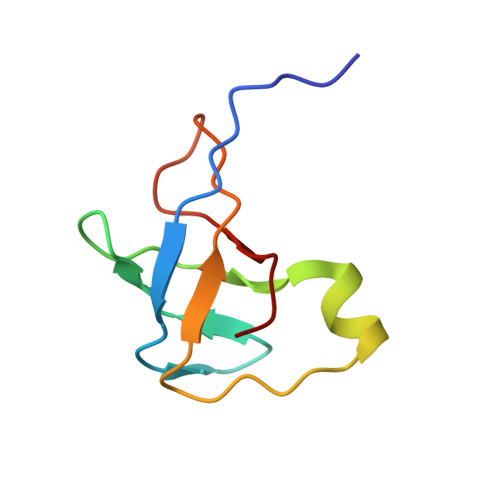
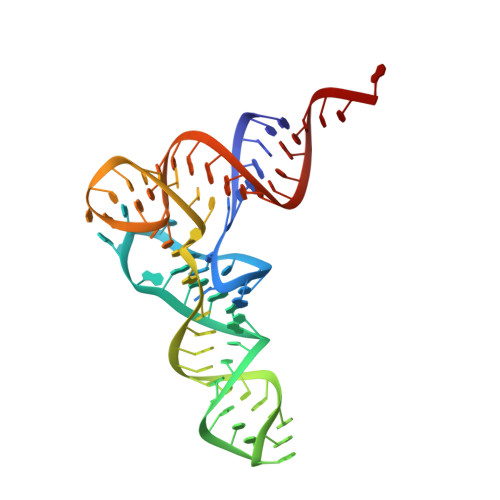
The Cryo-EM Structure of a Translation Initiation Complex from Escherichia coli.
Allen, G.S., Zavialov, A., Gursky, R., Ehrenberg, M., Frank, J.(2005) Cell 121: 703-712
- PubMed: 15935757
- DOI: https://doi.org/10.1016/j.cell.2005.03.023
- Primary Citation of Related Structures:
1ZO1, 1ZO3 - PubMed Abstract:
The 70S ribosome and its complement of factors required for initiation of translation in E. coli were purified separately and reassembled in vitro with GDPNP, producing a stable initiation complex (IC) stalled after 70S assembly. We have obtained a cryo-EM reconstruction of the IC showing IF2*GDPNP at the intersubunit cleft of the 70S ribosome. IF2*GDPNP contacts the 30S and 50S subunits as well as fMet-tRNA(fMet). IF2 here adopts a conformation radically different from that seen in the recent crystal structure of IF2. The C-terminal domain of IF2 binds to the single-stranded portion of fMet-tRNA(fMet), thereby forcing the tRNA into a novel orientation at the P site. The GTP binding domain of IF2 binds to the GTPase-associated center of the 50S subunit in a manner similar to EF-G and EF-Tu. Additionally, we present evidence for the localization of IF1, IF3, one C-terminal domain of L7/L12, and the N-terminal domain of IF2 in the initiation complex.
Organizational Affiliation:
Howard Hughes Medical Institute, Health Research, Inc. at the Wadsworth Center, Albany, New York 12201, USA.