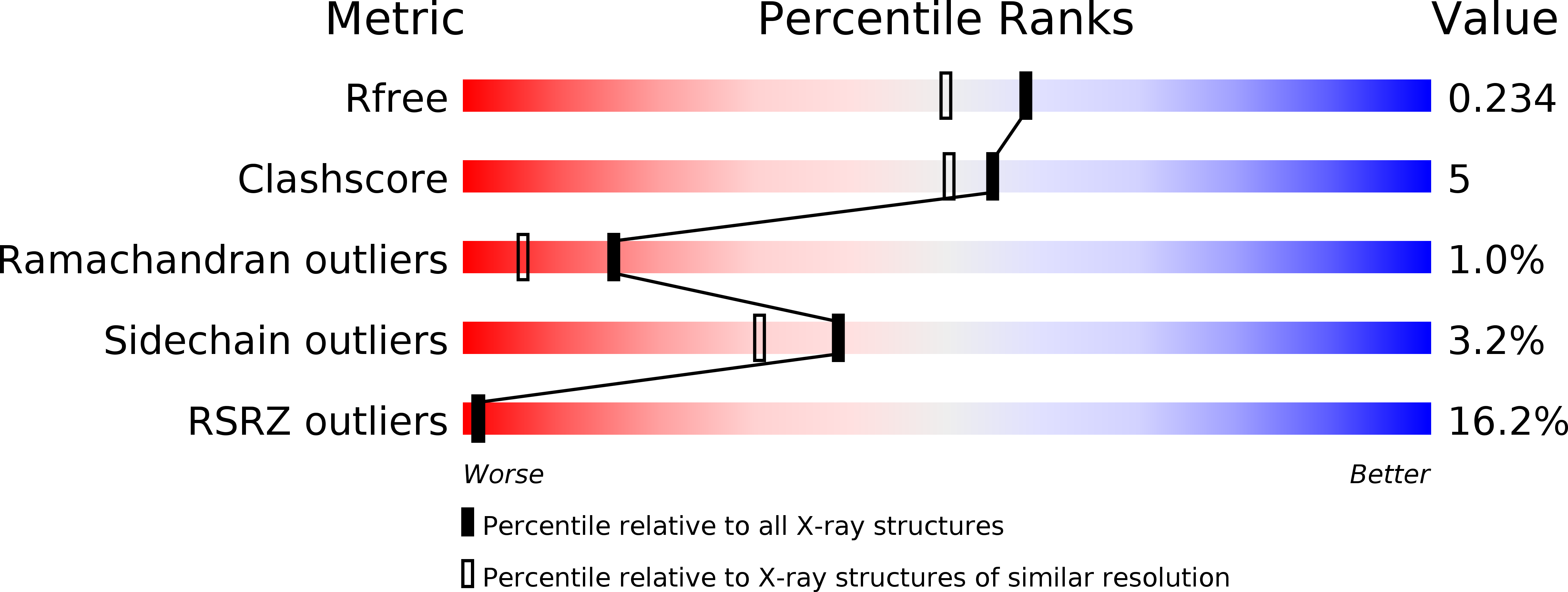
Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket
Bai, T., Becker, M., Gupta, A., Strike, P., Murphy, V.J., Anders, R.F., Batchelor, A.H.(2005) Proc Natl Acad Sci U S A 102: 12736-12741
- PubMed: 16129835
- DOI: https://doi.org/10.1073/pnas.0501808102
- Primary Citation of Related Structures:
1Z40 - PubMed Abstract:
Apical membrane antigen 1 (AMA1) is a leading malaria vaccine candidate that possesses polymorphisms that may pose a problem for a vaccine based on this antigen. Knowledge of the distribution of the polymorphic sites on the surface of AMA1 is necessary to obtain a detailed understanding of their significance for vaccine development. For this reason we have sought to determine the three-dimensional structure of AMA1 using x-ray crystallography. The central two-thirds of AMA1 is relatively conserved among Plasmodium species as well as more distantly related apicomplexan parasites, and contains two clusters of disulfide-bonded cysteines termed domains I and II. The crystal structure of this fragment of AMA1 reported here reveals that domains I+II consists of two intimately associated PAN domains. PAN domain I contains many long loops that extend from the domain core and form a scaffold for numerous polymorphic residues. This extreme adaptation of a PAN domain reveals how malaria parasites have introduced significant flexibility and variation into AMA1 to evade protective human antibody responses. The polymorphisms on the AMA1 surface are exclusively located on one side of the molecule, presumably because this region of AMA1 is most accessible to antibodies reacting with the parasite surface. Moreover, the most highly polymorphic residues surround a conserved hydrophobic trough that is ringed by domain I and domain II loops. Precedents set by viral receptor proteins would suggest that this is likely to be the AMA1 receptor binding pocket.
Organizational Affiliation:
University of Maryland School of Pharmacy, 20 Penn Street, Baltimore, MD 21201, USA.