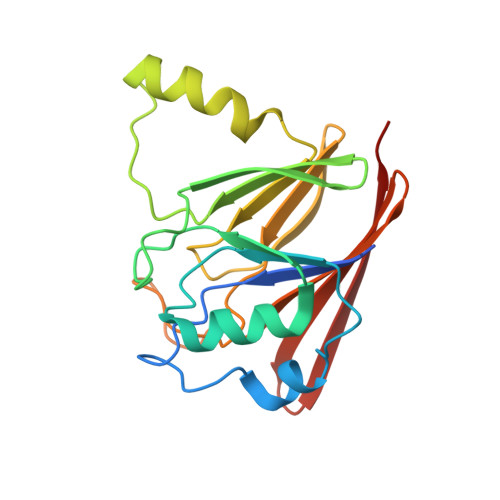
Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly
Collins, B.M., Skinner, C.F., Watson, P.J., Seaman, M.N.J., Owen, D.J.(2005) Nat Struct Mol Biol 12: 594-602
- PubMed: 15965486
- DOI: https://doi.org/10.1038/nsmb954
- Primary Citation of Related Structures:
1Z2W, 1Z2X - PubMed Abstract:
The retromer complex is responsible for the retrieval of mannose 6-phosphate receptors from the endosomal system to the Golgi. Here we present the crystal structure of the mammalian retromer subunit mVps29 and show that it has structural similarity to divalent metal-containing phosphoesterases. mVps29 can coordinate metals in a similar manner but has no detectable phosphoesterase activity in vitro, suggesting a unique specificity or function. The mVps29 and mVps26 subunits bind independently to mVps35 and together form a high-affinity heterotrimeric subcomplex. Mutagenesis reveals the structural basis for the interaction of mVps29 with mVps35 and subsequent association with endosomal membranes in vivo. A conserved hydrophobic surface distinct from the primary Vps35p binding site mediates assembly of the Vps29p-Vps26p-Vps35p subcomplex with sorting nexins in yeast, and mutation of either site results in a defect in retromer-dependent membrane trafficking.
Organizational Affiliation:
Cambridge Institute for Medical Research, Department of Clinical Biochemistry, University of Cambridge, Hills Road, Cambridge CB2 2XY, UK.